Faecal microbiota characterisation of horses using 16 rdna barcoded pyrosequencing, and carriage rate of clostridium difficile at hospital admission
- PMID: 26377067
- PMCID: PMC4573688
- DOI: 10.1186/s12866-015-0514-5
Faecal microbiota characterisation of horses using 16 rdna barcoded pyrosequencing, and carriage rate of clostridium difficile at hospital admission
Abstract
Background: The equine faecal microbiota is very complex and remains largely unknown, while interspecies interactions have an important contribution to animal health. Clostridium difficile has been identified as an important cause of diarrhoea in horses. This study provides further information on the nature of the bacterial communities present in horses developing an episode of diarrhoea. The prevalence of C. difficile in hospitalised horses at the time of admission is also reported.
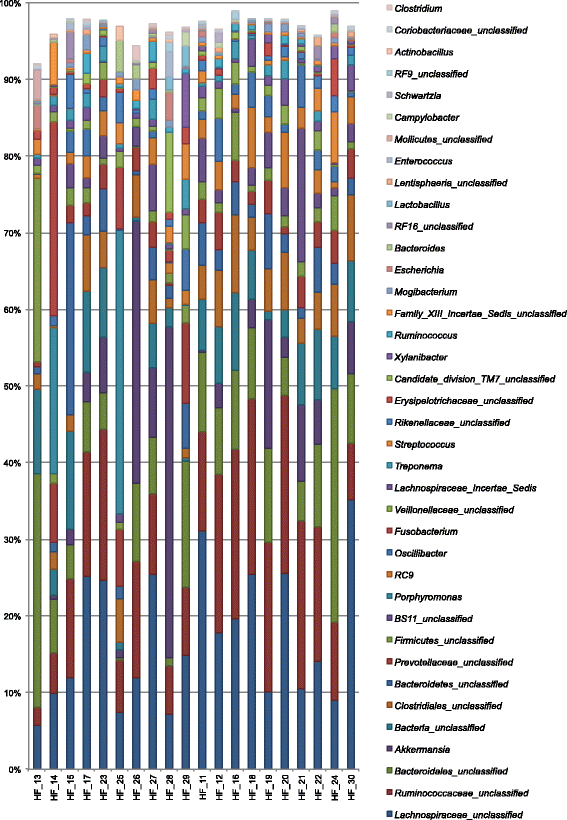
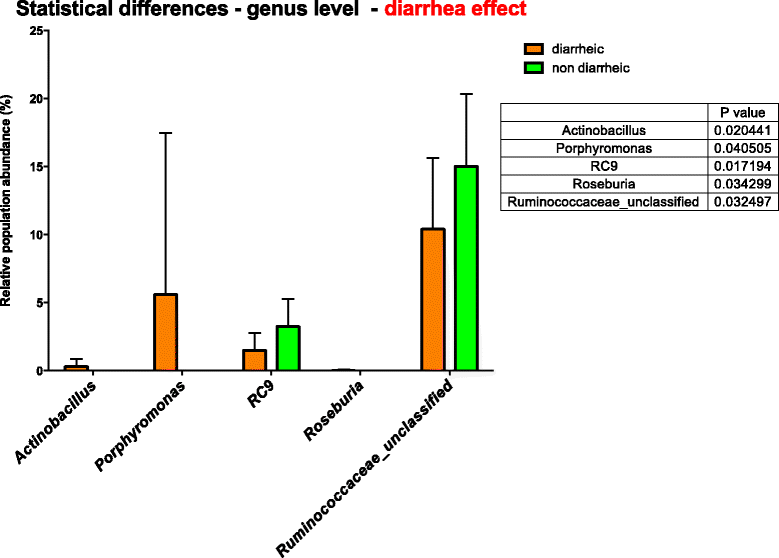
Results: Bacterial diversity of the gut microbiota in diarrhoea is lower than that in non-diarrhoeic horses in terms of species richness (p-value <0.002) and in population evenness (p-value: 0.02). Statistical differences for Actinobacillus, Porphyromonas, RC9 group, Roseburia and Ruminococcaceae were revealed. Fusobacteria was found in horses with diarrhoea but not in any of the horses with non-diarrheic faeces. In contrast, Akkermansia was among the three predominant taxa in all of the horses studied. The overall prevalence of C. difficile in the total samples of hospitalised horses at admission was 3.7 % (5/134), with five different PCR-ribotypes identified, including PCR-ribotype 014. Two colonised horses displayed a decreased bacterial species richness compared to the remaining subjects studied, which shared the same Bacteroides genus. However, none of the positive animals had diarrhoea at the moment of sampling.
Conclusions: The abundance of some taxa in the faecal microbiota of diarrhoeic horses can be a result of microbiome dysbiosis, and therefore a cause of intestinal disease, or some of these taxa may act as equine enteric pathogens. Clostridium difficile colonisation seems to be transient in all of the horses studied, without overgrowth to trigger infection. A large proportion of the sequences were unclassified, showing the complexity of horses' faecal microbiota.
Figures



References
-
- Daly K, Stewart CS, Flint HJ, Shirazi-Beechey SP. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiology Ecology. 2001;38:141–151. doi: 10.1111/j.1574-6941.2001.tb00892.x. - DOI
-
- Niwa C, Kato H, Hobo S, Kinoshita Y, Veno T, Katayama Y, et al. Postoperative Clostridium difficile infection with PCR ribotype 078 strain identified at necropsy in five throughbred racehorses. Veterinary Research. 2013;173:607. - PubMed
-
- Donaldson MT, Palmer JE. Prevalence of Clostridium perfringens enterotoxin and Clostridium difficile toxin A in feces of horses with diarrhea and colic. Journal of the American Veterinary Medical Association. 1999;215:358–61. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

