Differential regulation of metabolic pathways by androgen receptor (AR) and its constitutively active splice variant, AR-V7, in prostate cancer cells
- PMID: 26378018
- PMCID: PMC4741655
- DOI: 10.18632/oncotarget.5585
Differential regulation of metabolic pathways by androgen receptor (AR) and its constitutively active splice variant, AR-V7, in prostate cancer cells
Abstract
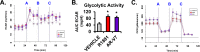
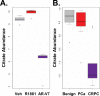
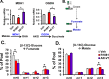
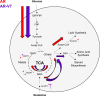
Metastatic prostate cancer (PCa) is primarily an androgen-dependent disease, which is treated with androgen deprivation therapy (ADT). Tumors usually develop resistance (castration-resistant PCa [CRPC]), but remain androgen receptor (AR) dependent. Numerous mechanisms for AR-dependent resistance have been identified including expression of constitutively active AR splice variants lacking the hormone-binding domain. Recent clinical studies show that expression of the best-characterized AR variant, AR-V7, correlates with resistance to ADT and poor outcome. Whether AR-V7 is simply a constitutively active substitute for AR or has novel gene targets that cause unique downstream changes is unresolved. Several studies have shown that AR activation alters cell metabolism. Using LNCaP cells with inducible expression of AR-V7 as a model system, we found that AR-V7 stimulated growth, migration, and glycolysis measured by ECAR (extracellular acidification rate) similar to AR. However, further analyses using metabolomics and metabolic flux assays revealed several differences. Whereas AR increased citrate levels, AR-V7 reduced citrate mirroring metabolic shifts observed in CRPC patients. Flux analyses indicate that the low citrate is a result of enhanced utilization rather than a failure to synthesize citrate. Moreover, flux assays suggested that compared to AR, AR-V7 exhibits increased dependence on glutaminolysis and reductive carboxylation to produce some of the TCA (tricarboxylic acid cycle) metabolites. These findings suggest that these unique actions represent potential therapeutic targets.
Keywords: LNCaP; androgen receptor; metabolism; prostate cancer; splice variant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–38. - PubMed
-
- Gelmann EP. Molecular biology of the androgen receptor. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2002;20:3001–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

