Evidence for an Ancestral Association of Human Coronavirus 229E with Bats
- PMID: 26378164
- PMCID: PMC4645311
- DOI: 10.1128/JVI.01755-15
Evidence for an Ancestral Association of Human Coronavirus 229E with Bats
Abstract
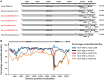
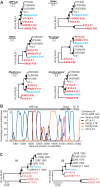
We previously showed that close relatives of human coronavirus 229E (HCoV-229E) exist in African bats. The small sample and limited genomic characterizations have prevented further analyses so far. Here, we tested 2,087 fecal specimens from 11 bat species sampled in Ghana for HCoV-229E-related viruses by reverse transcription-PCR (RT-PCR). Only hipposiderid bats tested positive. To compare the genetic diversity of bat viruses and HCoV-229E, we tested historical isolates and diagnostic specimens sampled globally over 10 years. Bat viruses were 5- and 6-fold more diversified than HCoV-229E in the RNA-dependent RNA polymerase (RdRp) and spike genes. In phylogenetic analyses, HCoV-229E strains were monophyletic and not intermixed with animal viruses. Bat viruses formed three large clades in close and more distant sister relationships. A recently described 229E-related alpaca virus occupied an intermediate phylogenetic position between bat and human viruses. According to taxonomic criteria, human, alpaca, and bat viruses form a single CoV species showing evidence for multiple recombination events. HCoV-229E and the alpaca virus showed a major deletion in the spike S1 region compared to all bat viruses. Analyses of four full genomes from 229E-related bat CoVs revealed an eighth open reading frame (ORF8) located at the genomic 3' end. ORF8 also existed in the 229E-related alpaca virus. Reanalysis of HCoV-229E sequences showed a conserved transcription regulatory sequence preceding remnants of this ORF, suggesting its loss after acquisition of a 229E-related CoV by humans. These data suggested an evolutionary origin of 229E-related CoVs in hipposiderid bats, hypothetically with camelids as intermediate hosts preceding the establishment of HCoV-229E.
Importance: The ancestral origins of major human coronaviruses (HCoVs) likely involve bat hosts. Here, we provide conclusive genetic evidence for an evolutionary origin of the common cold virus HCoV-229E in hipposiderid bats by analyzing a large sample of African bats and characterizing several bat viruses on a full-genome level. Our evolutionary analyses show that animal and human viruses are genetically closely related, can exchange genetic material, and form a single viral species. We show that the putative host switches leading to the formation of HCoV-229E were accompanied by major genomic changes, including deletions in the viral spike glycoprotein gene and loss of an open reading frame. We reanalyze a previously described genetically related alpaca virus and discuss the role of camelids as potential intermediate hosts between bat and human viruses. The evolutionary history of HCoV-229E likely shares important characteristics with that of the recently emerged highly pathogenic Middle East respiratory syndrome (MERS) coronavirus.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, Woo PCY, Ziebuhr J. 2012. Family Coronaviridae, p 806–820. In King A, Adams M, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, Amsterdam, The Netherlands.
-
- van der Hoek L. 2007. Human coronaviruses: what do they cause? Antivir Ther 12:651–658. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

