Mitigating false-positive associations in rare disease gene discovery
- PMID: 26378430
- PMCID: PMC4576452
- DOI: 10.1002/humu.22847
Mitigating false-positive associations in rare disease gene discovery
Abstract
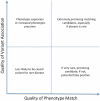
Clinical sequencing is expanding, but causal variants are still not identified in the majority of cases. These unsolved cases can aid in gene discovery when individuals with similar phenotypes are identified in systems such as the Matchmaker Exchange. We describe risks for gene discovery in this growing set of unsolved cases. In a set of rare disease cases with the same phenotype, it is not difficult to find two individuals with the same phenotype that carry variants in the same gene. We quantify the risk of false-positive association in a cohort of individuals with the same phenotype, using the prior probability of observing a variant in each gene from over 60,000 individuals (Exome Aggregation Consortium). Based on the number of individuals with a genic variant, cohort size, specific gene, and mode of inheritance, we calculate a P value that the match represents a true association. A match in two of 10 patients in MECP2 is statistically significant (P = 0.0014), whereas a match in TTN would not reach significance, as expected (P > 0.999). Finally, we analyze the probability of matching in clinical exome cases to estimate the number of cases needed to identify genes related to different disorders. We offer Rare Disease Match, an online tool to mitigate the uncertainty of false-positive associations.
Keywords: Matchmaker Exchange; false-positive associations; incidental findings; incidentalome; matchmaking; rare diseases.
© 2015 WILEY PERIODICALS, INC.
Figures

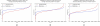
References
-
- 2014. (ExAC) EAC.
-
- Brownstein CA, Beggs AH, Homer N, Merriman B, Yu TW, Flannery KC, Dechene ET, Towne MC, Savage SK, Price EN, Holm IA, Luquette LJ, et al. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome Biol. 2014;15:R53. - PMC - PubMed
-
- Collection S. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014:1–95. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

