The Role of ARF6 in Biliary Atresia
- PMID: 26379158
- PMCID: PMC4574480
- DOI: 10.1371/journal.pone.0138381
The Role of ARF6 in Biliary Atresia
Abstract
Background & aims: Altered extrahepatic bile ducts, gut, and cardiovascular anomalies constitute the variable phenotype of biliary atresia (BA).
Methods: To identify potential susceptibility loci, Caucasian children, normal (controls) and with BA (cases) at two US centers were compared at >550000 SNP loci. Systems biology analysis was carried out on the data. In order to validate a key gene identified in the analysis, biliary morphogenesis was evaluated in 2-5-day post-fertilization zebrafish embryos after morpholino-antisense oligonucleotide knockdown of the candidate gene ADP ribosylation factor-6 (ARF6, Mo-arf6).
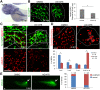
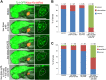
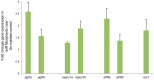
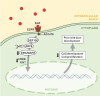
Results: Among 39 and 24 cases at centers 1 and 2, respectively, and 1907 controls, which clustered together on principal component analysis, the SNPs rs3126184 and rs10140366 in a 3' flanking enhancer region for ARF6 demonstrated higher minor allele frequencies (MAF) in each cohort, and 63 combined cases, compared with controls (0.286 vs. 0.131, P = 5.94x10-7, OR 2.66; 0.286 vs. 0.13, P = 5.57x10-7, OR 2.66). Significance was enhanced in 77 total cases, which included 14 additional BA genotyped at rs3126184 only (p = 1.58x10-2, OR = 2.66). Pathway analysis of the 1000 top-ranked SNPs in CHP cases revealed enrichment of genes for EGF regulators (p<1 x10-7), ERK/MAPK and CREB canonical pathways (p<1 x10-34), and functional networks for cellular development and proliferation (p<1 x10-45), further supporting the role of EGFR-ARF6 signaling in BA. In zebrafish embryos, Mo-arf6 injection resulted in a sparse intrahepatic biliary network, several biliary epithelial cell defects, and poor bile excretion to the gall bladder compared with uninjected embryos. Biliary defects were reproduced with the EGFR-blocker AG1478 alone or with Mo-arf6 at lower doses of each agent and rescued with arf6 mRNA.
Conclusions: The BA-associated SNPs identify a chromosome 14q21.3 susceptibility locus encompassing the ARF6 gene. arf6 knockdown in zebrafish implicates early biliary dysgenesis as a basis for BA, and also suggests a role for EGFR signaling in BA pathogenesis.
Conflict of interest statement
Figures



References
-
- Karrer FM, Price MR, Bensard DD, Sokol RJ, Narkewicz MR, Smith DJ, et al. (1996) Long-term results with the Kasai operation for biliary atresia. Arch Surg 131: 493–496. - PubMed
-
- Russo P, Magee JC, Boitnott J, Bove KE, Raghunathan T, Finegold M, et al. (2011) Biliary Atresia Research Consortium. Design and validation of the biliary atresia research consortium histologic assessment system for cholestasis in infancy. Clin Gastroenterol Hepatol 9: 357–362e2. 10.1016/j.cgh.2011.01.003 - DOI - PMC - PubMed
-
- Desmet VJ. (1998) Ludwig symposium on biliary disorders—part I. Pathogenesis of ductal plate abnormalities. Mayo Clin Proc 73: 80–89. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

