EEGNET: An Open Source Tool for Analyzing and Visualizing M/EEG Connectome
- PMID: 26379232
- PMCID: PMC4574940
- DOI: 10.1371/journal.pone.0138297
EEGNET: An Open Source Tool for Analyzing and Visualizing M/EEG Connectome
Abstract
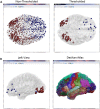
The brain is a large-scale complex network often referred to as the "connectome". Exploring the dynamic behavior of the connectome is a challenging issue as both excellent time and space resolution is required. In this context Magneto/Electroencephalography (M/EEG) are effective neuroimaging techniques allowing for analysis of the dynamics of functional brain networks at scalp level and/or at reconstructed sources. However, a tool that can cover all the processing steps of identifying brain networks from M/EEG data is still missing. In this paper, we report a novel software package, called EEGNET, running under MATLAB (Math works, inc), and allowing for analysis and visualization of functional brain networks from M/EEG recordings. EEGNET is developed to analyze networks either at the level of scalp electrodes or at the level of reconstructed cortical sources. It includes i) Basic steps in preprocessing M/EEG signals, ii) the solution of the inverse problem to localize / reconstruct the cortical sources, iii) the computation of functional connectivity among signals collected at surface electrodes or/and time courses of reconstructed sources and iv) the computation of the network measures based on graph theory analysis. EEGNET is the unique tool that combines the M/EEG functional connectivity analysis and the computation of network measures derived from the graph theory. The first version of EEGNET is easy to use, flexible and user friendly. EEGNET is an open source tool and can be freely downloaded from this webpage: https://sites.google.com/site/eegnetworks/.
Conflict of interest statement
Figures




References
-
- Bartolomei F, Bosma I, Klein M, Baayen JC, Reijneveld JC, Postma TJ, et al. How do brain tumors alter functional connectivity? A magnetoencephalography study. Annals of neurology. 2006;59(1):128–38. - PubMed
-
- Rodriguez E, George N, Lachaux JP, Martinerie J, Renault B, Varela FJ. Perception's shadow: long-distance synchronization of human brain activity. Nature. 1999;397(6718):430–3. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

