The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo
- PMID: 26379282
- PMCID: PMC4575032
- DOI: 10.1371/journal.ppat.1005142
The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo
Abstract
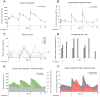
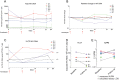
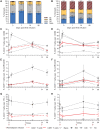
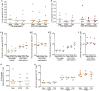
Pharmacologically-induced activation of replication competent proviruses from latency in the presence of antiretroviral treatment (ART) has been proposed as a step towards curing HIV-1 infection. However, until now, approaches to reverse HIV-1 latency in humans have yielded mixed results. Here, we report a proof-of-concept phase Ib/IIa trial where 6 aviremic HIV-1 infected adults received intravenous 5 mg/m2 romidepsin (Celgene) once weekly for 3 weeks while maintaining ART. Lymphocyte histone H3 acetylation, a cellular measure of the pharmacodynamic response to romidepsin, increased rapidly (maximum fold range: 3.7–7.7 relative to baseline) within the first hours following each romidepsin administration. Concurrently, HIV-1 transcription quantified as copies of cell-associated un-spliced HIV-1 RNA increased significantly from baseline during treatment (range of fold-increase: 2.4–5.0; p = 0.03). Plasma HIV-1 RNA increased from <20 copies/mL at baseline to readily quantifiable levels at multiple post-infusion time-points in 5 of 6 patients (range 46–103 copies/mL following the second infusion, p = 0.04). Importantly, romidepsin did not decrease the number of HIV-specific T cells or inhibit T cell cytokine production. Adverse events (all grade 1–2) were consistent with the known side effects of romidepsin. In conclusion, romidepsin safely induced HIV-1 transcription resulting in plasma HIV-1 RNA that was readily detected with standard commercial assays demonstrating that significant reversal of HIV-1 latency in vivo is possible without blunting T cell-mediated immune responses. These finding have major implications for future trials aiming to eradicate the HIV-1 reservoir.
Trial registration: clinicaltrials.gov NTC02092116.
Trial registration: ClinicalTrials.gov NCT02092116.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: MS is an employee of Bionor Pharma ASA and has shares in the company. KK is a consultant to Bionor Pharma ASA. The other authors declare no competing interests. This does not alter our adherence to all PLOS policies on sharing data and materials.
Figures

Comment in
-
Romidepsin reverses HIV-1 latency in vivo.AIDS. 2016 Mar 13;30(5):N3. doi: 10.1097/QAD.0000000000000998. AIDS. 2016. PMID: 26645606 No abstract available.
References
-
- Li TS, Tubiana R, Katlama C, Calvez V, Ait Mohand H, Autran B. Long-lasting recovery in CD4 T-cell function and viral-load reduction after highly active antiretroviral therapy in advanced HIV-1 disease. Lancet. 1998;351(9117):1682–6. Epub 1998/09/12. S0140673697102914 [pii]. . - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–300. Epub 1997/11/21. . - PubMed

