Combination of RNAseq and SNP nanofluidic array reveals the center of genetic diversity of cacao pathogen Moniliophthora roreri in the upper Magdalena Valley of Colombia and its clonality
- PMID: 26379633
- PMCID: PMC4550789
- DOI: 10.3389/fmicb.2015.00850
Combination of RNAseq and SNP nanofluidic array reveals the center of genetic diversity of cacao pathogen Moniliophthora roreri in the upper Magdalena Valley of Colombia and its clonality
Abstract
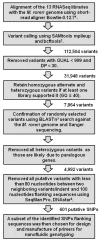
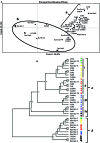
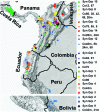
Moniliophthora roreri is the fungal pathogen that causes frosty pod rot (FPR) disease of Theobroma cacao L., the source of chocolate. FPR occurs in most of the cacao producing countries in the Western Hemisphere, causing yield losses up to 80%. Genetic diversity within the FPR pathogen population may allow the population to adapt to changing environmental conditions and adapt to enhanced resistance in the host plant. The present study developed single nucleotide polymorphism (SNP) markers from RNASeq results for 13 M. roreri isolates and validated the markers for their ability to reveal genetic diversity in an international M. roreri collection. The SNP resources reported herein represent the first study of RNA sequencing (RNASeq)-derived SNP validation in M. roreri and demonstrates the utility of RNASeq as an approach for de novo SNP identification in M. roreri. A total of 88 polymorphic SNPs were used to evaluate the genetic diversity of 172 M. roreri cacao isolates resulting in 37 distinct genotypes (including 14 synonymous groups). Absence of heterozygosity for the 88 SNP markers indicates reproduction in M. roreri is clonal and likely due to a homothallic life style. The upper Magdalena Valley of Colombia showed the highest levels of genetic diversity with 20 distinct genotypes of which 13 were limited to this region, and indicates this region as the possible center of origin for M. roreri.
Keywords: RNAseq; SNP; biodiversity; genotyping; homothallism; monilia pod rot.
Figures

References
-
- Ali S. S., Melnick R. L., Crozier J., Phillips-Mora W., Strem M. D., Shao J., et al. (2014). Successful pod infections by Moniliophthora roreri result in differential Theobroma cacao gene expression depending on the clone’s level of tolerance. Mol. Plant Pathol. 15 698–710. 10.1111/mpp.12126 - DOI - PMC - PubMed
-
- Ancízar M. (1853). Peregrinacion de Alpha: Por Las Provincias del Norte de la Nueva Granada, en 1850-51. Bogotá: Imprenta de Echeverría Hermanos.
-
- Bailey B. A., Crozier J., Sicher R. C., Strem M. D., Melnick R., Carazzolle M. F., et al. (2013). Dynamic changes in pod and fungal physiology associated with the shift from biotrophy to necrotrophy during the infection of Theobroma cacao by Moniliophthora roreri. Physiol. Mol. Plant Pathol. 81 84–96. 10.1016/j.pmpp.2012.11.005 - DOI
-
- Bailey B. A., Strem M. D., Bae H., de Mayolo G. A., Guiltinan M. J. (2005). Gene expression in leaves of Theobroma cacao in response to mechanical wounding, ethylene, and/or methyl jasmonate. Plant Sci. 168 1247–1258. 10.1016/j.plantsci.2005.01.002 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

