Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains
- PMID: 26379646
- PMCID: PMC4548156
- DOI: 10.3389/fmicb.2015.00871
Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains
Abstract
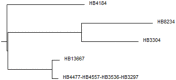
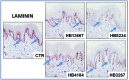
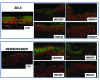
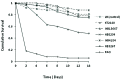
Pseudomonas putida strains are ubiquitous in soil and water but have also been reported as opportunistic human pathogens capable of causing nosocomial infections. In this study we describe the multilocus sequence typing of four P. putida strains (HB13667, HB8234, HB4184, and HB3267) isolated from in-patients at the Besançon Hospital (France). The four isolates (in particular HB3267) were resistant to a number of antibiotics. The pathogenicity and virulence potential of the strains was tested ex vivo and in vivo using different biological models: human tissue culture, mammalian tissues, and insect larvae. Our results showed a significant variability in the ability of the four strains to damage the host; HB13667 did not exhibit any pathogenic traits, HB4184 caused damage only ex vivo in human tissue cultures, and HB8234 had a deleterious effect in tissue culture and in vivo on rat skin, but not in insect larvae. Interestingly, strain HB3267 caused damage in all the model systems studied. The putative evolution of these strains in medical environments is discussed.
Keywords: Pseudomonas; hospitalary strains; laminin; opportunistic pathogen.
Figures



References
-
- Abdallah M., Benoliel C., Drider D., Dhulster P., Chihib N. E. (2014).Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 196 453–472. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

