Toward a standard in structural genome annotation for prokaryotes
- PMID: 26380633
- PMCID: PMC4572445
- DOI: 10.1186/s40793-015-0034-9
Toward a standard in structural genome annotation for prokaryotes
Abstract
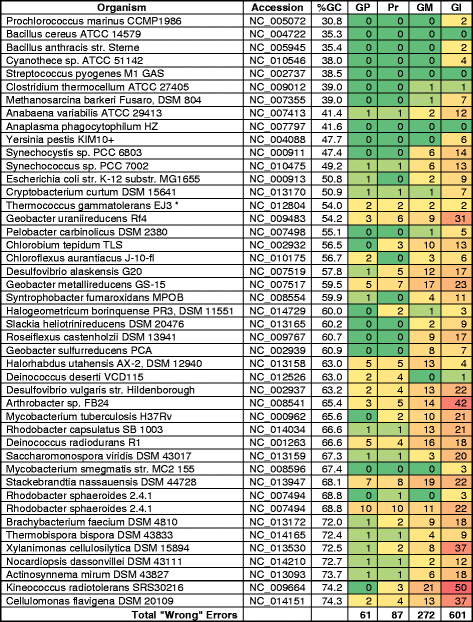
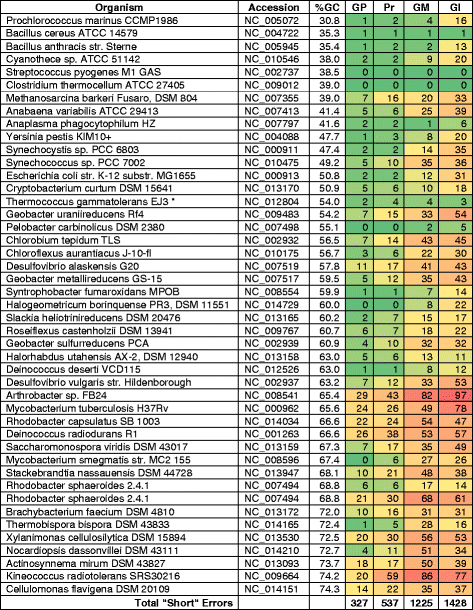
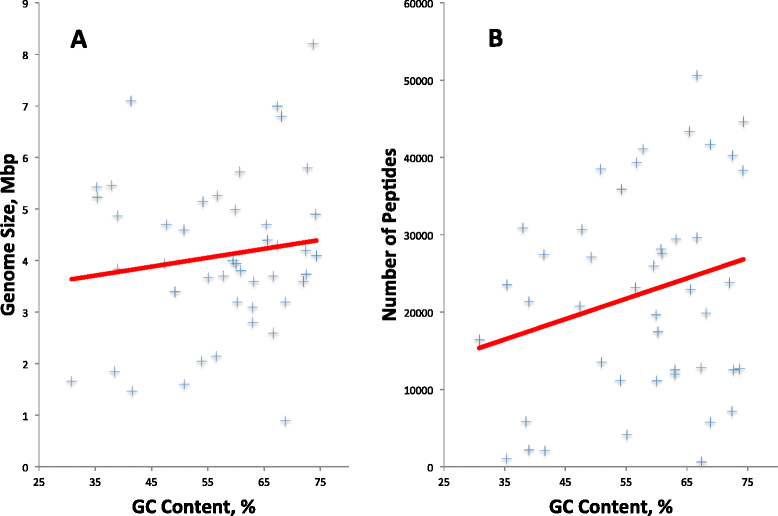
Background: In an effort to identify the best practice for finding genes in prokaryotic genomes and propose it as a standard for automated annotation pipelines, 1,004,576 peptides were collected from various publicly available resources, and were used as a basis to evaluate various gene-calling methods. The peptides came from 45 bacterial replicons with an average GC content from 31 % to 74 %, biased toward higher GC content genomes. Automated, manual, and semi-manual methods were used to tally errors in three widely used gene calling methods, as evidenced by peptides mapped outside the boundaries of called genes.
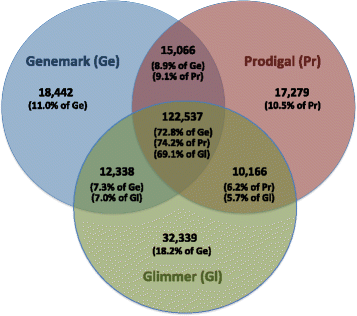
Results: We found that the consensus set of identical genes predicted by the three methods constitutes only about 70 % of the genes predicted by each individual method (with start and stop required to coincide). Peptide data was useful for evaluating some of the differences between gene callers, but not reliable enough to make the results conclusive, due to limitations inherent in any proteogenomic study.
Conclusions: A single, unambiguous, unanimous best practice did not emerge from this analysis, since the available proteomics data were not adequate to provide an objective measurement of differences in the accuracy between these methods. However, as a result of this study, software, reference data, and procedures have been better matched among participants, representing a step toward a much-needed standard. In the absence of sufficient amount of exprimental data to achieve a universal standard, our recommendation is that any of these methods can be used by the community, as long as a single method is employed across all datasets to be compared.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

