An ensemble approach to accurately detect somatic mutations using SomaticSeq
- PMID: 26381235
- PMCID: PMC4574535
- DOI: 10.1186/s13059-015-0758-2
An ensemble approach to accurately detect somatic mutations using SomaticSeq
Abstract
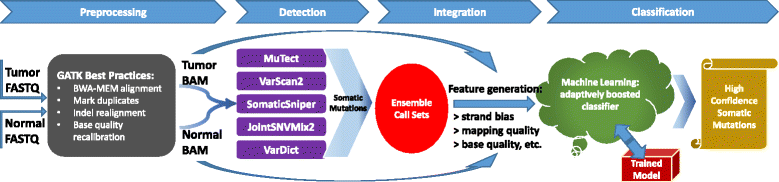
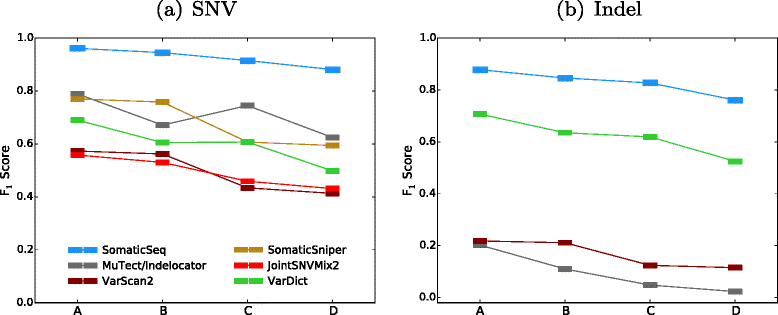
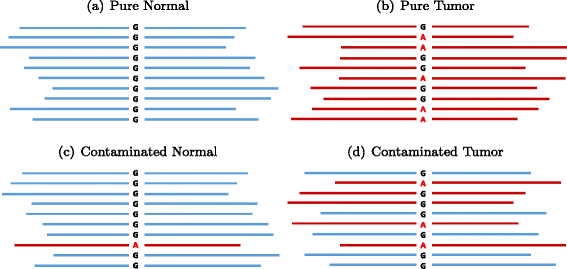
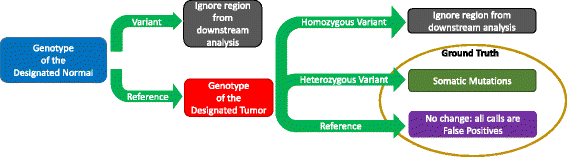
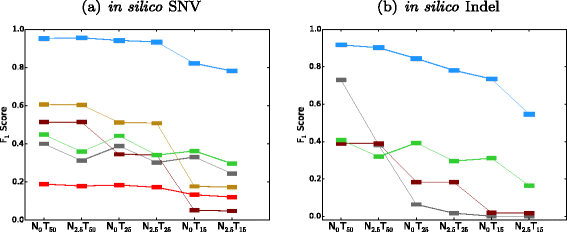
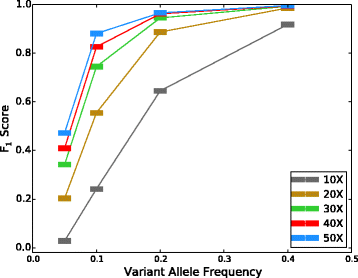
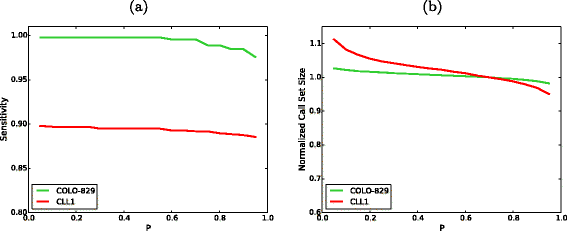
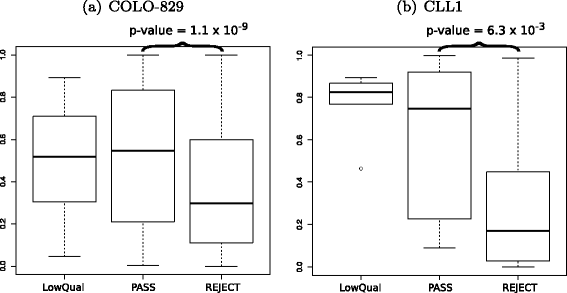
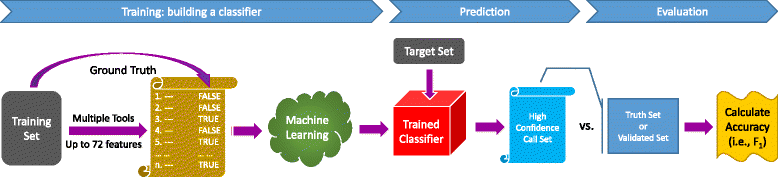
SomaticSeq is an accurate somatic mutation detection pipeline implementing a stochastic boosting algorithm to produce highly accurate somatic mutation calls for both single nucleotide variants and small insertions and deletions. The workflow currently incorporates five state-of-the-art somatic mutation callers, and extracts over 70 individual genomic and sequencing features for each candidate site. A training set is provided to an adaptively boosted decision tree learner to create a classifier for predicting mutation statuses. We validate our results with both synthetic and real data. We report that SomaticSeq is able to achieve better overall accuracy than any individual tool incorporated.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

