Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy
- PMID: 26381381
- PMCID: PMC4660980
- DOI: 10.4155/fmc.15.101
Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy
Abstract
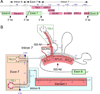
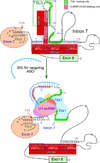
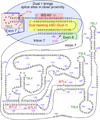
Spinal muscular atrophy (SMA) is a major neurodegenerative disorder of children and infants. SMA is primarily caused by low levels of SMN protein owing to deletions or mutations of the SMN1 gene. SMN2, a nearly identical copy of SMN1, fails to compensate for the loss of the production of the functional SMN protein due to predominant skipping of exon 7. Several compounds, including antisense oligonucleotides (ASOs) that elevate SMN protein from SMN2 hold the promise for treatment. An ASO-based drug currently under Phase III clinical trial employs intronic splicing silencer N1 (ISS-N1) as its target. Cumulative studies on ISS-N1 reveal a wealth of information with significance to the overall therapeutic development for SMA. Here, the authors summarize the mechanistic principles behind various antisense targets currently available for SMA therapy.
Figures

References
-
- Prior TW. Spinal muscular atrophy diagnostics. J. Child Neurol. 2007;22(8):952–956. - PubMed
-
- Tiziano FD, Melki J, Simard LR. Solving the puzzle of spinal muscular atrophy: what are the missing pieces? Am. J. Med. Genet. A. 2013;161A(11):2836–2845. - PubMed
-
- Nurputra DK, Lai PS, Harahap NIF, et al. Spinal Muscular Atrophy: From Gene Discovery to Clinical Trials. Ann. Hum. Genet. 2013;77(5):435–463. - PubMed
-
- Lefebvre S, Bürglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
