Multiple structural and epigenetic defects in the human leukocyte antigen class I antigen presentation pathway in a recurrent metastatic melanoma following immunotherapy
- PMID: 26381407
- PMCID: PMC4646314
- DOI: 10.1074/jbc.M115.676130
Multiple structural and epigenetic defects in the human leukocyte antigen class I antigen presentation pathway in a recurrent metastatic melanoma following immunotherapy
Abstract
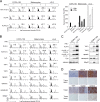
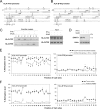
Scant information is available about the molecular basis of multiple HLA class I antigen-processing machinery defects in malignant cells, although this information contributes to our understanding of the molecular immunoescape mechanisms utilized by tumor cells and may suggest strategies to counteract them. In the present study we reveal a combination of IFN-γ-irreversible structural and epigenetic defects in HLA class I antigen-processing machinery in a recurrent melanoma metastasis after immunotherapy. These defects include loss of tapasin and one HLA haplotype as well as selective silencing of HLA-A3 gene responsiveness to IFN-γ. Tapasin loss is caused by a germ-line frameshift mutation in exon 3 (TAPBP(684delA)) along with a somatic loss of the other gene copy. Selective silencing of HLA-A3 gene and its IFN-γ responsiveness is associated with promoter CpG methylation nearby site-α and TATA box, reversible after DNA methyltransferase 1 depletion. This treatment combined with tapasin reconstitution and IFN-γ stimulation restored the highest level of HLA class I expression and its ability to elicit cytotoxic T cell responses. These results represent a novel tumor immune evasion mechanism through impairing multiple components at various levels in the HLA class I antigen presentation pathway. These findings may suggest a rational design of combinatorial cancer immunotherapy harnessing DNA demethylation and IFN-γ response.
Keywords: DNA methylation; antigen presentation; immunotherapy; major histocompatibility complex (MHC); tumor immunology.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Hodi F. S., O'Day S. J., McDermott D. F., Weber R. W., Sosman J. A., Haanen J. B., Gonzalez R., Robert C., Schadendorf D., Hassel J. C., Akerley W., van den Eertwegh A. J., Lutzky J., Lorigan P., Vaubel J. M., Linette G. P., Hogg D., Ottensmeier C. H., Lebbé C., Peschel C., Quirt I., Clark J. I., Wolchok J. D., Weber J. S., Tian J., Yellin M. J., Nichol G. M., Hoos A., and Urba W. J. (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 - PMC - PubMed
-
- Hamid O., Robert C., Daud A., Hodi F. S., Hwu W. J., Kefford R., Wolchok J. D., Hersey P., Joseph R. W., Weber J. S., Dronca R., Gangadhar T. C., Patnaik A., Zarour H., Joshua A. M., Gergich K., Elassaiss-Schaap J., Algazi A., Mateus C., Boasberg P., Tumeh P. C., Chmielowski B., Ebbinghaus S. W., Li X. N., Kang S. P., and Ribas A. (2013) Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144 - PMC - PubMed
-
- Sharma P., and Allison J. P. (2015) The future of immune checkpoint therapy. Science 348, 56–61 - PubMed
-
- Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J. M., Desrichard A., Walsh L. A., Postow M. A., Wong P., Ho T. S., Hollmann T. J., Bruggeman C., Kannan K., Li Y., Elipenahli C., Liu C., Harbison C. T., Wang L., Ribas A., Wolchok J. D., and Chan T. A. (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 - PMC - PubMed
-
- Rizvi N. A., Hellmann M. D., Snyder A., Kvistborg P., Makarov V., Havel J. J., Lee W., Yuan J., Wong P., Ho T. S., Miller M. L., Rekhtman N., Moreira A. L., Ibrahim F., Bruggeman C., Gasmi B., Zappasodi R., Maeda Y., Sander C., Garon E. B., Merghoub T., Wolchok J. D., Schumacher T. N., and Chan T. A. (2015) Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

