De novo peroxisome biogenesis: Evolving concepts and conundrums
- PMID: 26381541
- PMCID: PMC4791208
- DOI: 10.1016/j.bbamcr.2015.09.014
De novo peroxisome biogenesis: Evolving concepts and conundrums
Abstract
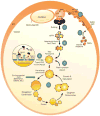
Peroxisomes proliferate by growth and division of pre-existing peroxisomes or could arise de novo. Though the de novo pathway of peroxisome biogenesis is a more recent discovery, several studies have highlighted key mechanistic details of the pathway. The endoplasmic reticulum (ER) is the primary source of lipids and proteins for the newly-formed peroxisomes. More recently, an intricate sorting process functioning at the ER has been proposed, that segregates specific PMPs first to peroxisome-specific ER domains (pER) and then assembles PMPs selectively into distinct pre-peroxisomal vesicles (ppVs) that later fuse to form import-competent peroxisomes. In addition, plausible roles of the three key peroxins Pex3, Pex16 and Pex19, which are also central to the growth and division pathway, have been suggested in the de novo process. In this review, we discuss key developments and highlight the unexplored avenues in de novo peroxisome biogenesis.
Keywords: De novo peroxisome biogenesis; Intra-ER sorting, import of PMPs into the ER, growth and division vs de novo peroxisome biogenesis; Pre-peroxisomal vesicles, ppVs; Pre-peroxisomal-ER, pER; Role of endoplasmic reticulum in peroxisome biogenesis, peroxisomal membrane protein biogenesis; Role of the endoplasmic reticulum in yeast, mammals and plants.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures

References
-
- Rhodin J. Correlation of Ultrastructural Organization and Function in Normal and Experimentally Changed Proximal Convoluted Tubule Cells of the Mouse Kidney: An Electron Microscopic Study. Including an Experimental Analysis of the Conditions for Fixation of the Renal Tissue for High Resolution Electron Microscopy. 1954;3:187–206.
-
- De Duve C. Intracellular localization of enzymes. Nature. 1960;187:836–853. doi: 10.1038/187836a0. - DOI
-
- De Duve C. Functions of microbodies (peroxisomes) J Cell Biol. 1965;27:25A–26A. - PubMed
-
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

