Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data
- PMID: 26383846
- PMCID: PMC4858396
- DOI: 10.1289/ehp.1509763
Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data
Abstract
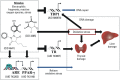
Background: Hepatotoxicity accounts for a substantial number of drugs being withdrawn from the market. Using traditional animal models to detect hepatotoxicity is expensive and time-consuming. Alternative in vitro methods, in particular cell-based high-throughput screening (HTS) studies, have provided the research community with a large amount of data from toxicity assays. Among the various assays used to screen potential toxicants is the antioxidant response element beta lactamase reporter gene assay (ARE-bla), which identifies chemicals that have the potential to induce oxidative stress and was used to test > 10,000 compounds from the Tox21 program.
Objective: The ARE-bla computational model and HTS data from a big data source (PubChem) were used to profile environmental and pharmaceutical compounds with hepatotoxicity data.
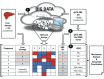
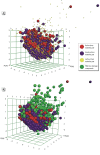
Methods: Quantitative structure-activity relationship (QSAR) models were developed based on ARE-bla data. The models predicted the potential oxidative stress response for known liver toxicants when no ARE-bla data were available. Liver toxicants were used as probe compounds to search PubChem Bioassay and generate a response profile, which contained thousands of bioassays (> 10 million data points). By ranking the in vitro-in vivo correlations (IVIVCs), the most relevant bioassay(s) related to hepatotoxicity were identified.
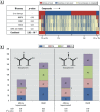
Results: The liver toxicants profile contained the ARE-bla and relevant PubChem assays. Potential toxicophores for well-known toxicants were created by identifying chemical features that existed only in compounds with high IVIVCs.
Conclusion: Profiling chemical IVIVCs created an opportunity to fully explore the source-to-outcome continuum of modern experimental toxicology using cheminformatics approaches and big data sources.
Citation: Kim MT, Huang R, Sedykh A, Wang W, Xia M, Zhu H. 2016. Mechanism profiling of hepatotoxicity caused by oxidative stress using antioxidant response element reporter gene assay models and big data. Environ Health Perspect 124:634-641; http://dx.doi.org/10.1289/ehp.1509763.
Conflict of interest statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors declare they have no actual or potential competing financial interests.
Figures
Similar articles
-
Profiling animal toxicants by automatically mining public bioassay data: a big data approach for computational toxicology.PLoS One. 2014 Jun 20;9(6):e99863. doi: 10.1371/journal.pone.0099863. eCollection 2014. PLoS One. 2014. PMID: 24950175 Free PMC article.
-
Computational toxicology as implemented by the U.S. EPA: providing high throughput decision support tools for screening and assessing chemical exposure, hazard and risk.J Toxicol Environ Health B Crit Rev. 2010 Feb;13(2-4):197-217. doi: 10.1080/10937404.2010.483935. J Toxicol Environ Health B Crit Rev. 2010. PMID: 20574897
-
Use of in vitro HTS-derived concentration-response data as biological descriptors improves the accuracy of QSAR models of in vivo toxicity.Environ Health Perspect. 2011 Mar;119(3):364-70. doi: 10.1289/ehp.1002476. Epub 2010 Oct 27. Environ Health Perspect. 2011. PMID: 20980217 Free PMC article.
-
Big data in chemical toxicity research: the use of high-throughput screening assays to identify potential toxicants.Chem Res Toxicol. 2014 Oct 20;27(10):1643-51. doi: 10.1021/tx500145h. Epub 2014 Sep 16. Chem Res Toxicol. 2014. PMID: 25195622 Free PMC article. Review.
-
The use of high-throughput screening techniques to evaluate mitochondrial toxicity.Toxicology. 2017 Nov 1;391:34-41. doi: 10.1016/j.tox.2017.07.020. Epub 2017 Aug 5. Toxicology. 2017. PMID: 28789971 Review.
Cited by
-
Supporting read-across using biological data.ALTEX. 2016;33(2):167-82. doi: 10.14573/altex.1601252. Epub 2016 Feb 11. ALTEX. 2016. PMID: 26863516 Free PMC article.
-
Predicting Chemical Immunotoxicity through Data-Driven QSAR Modeling of Aryl Hydrocarbon Receptor Agonism and Related Toxicity Mechanisms.Environ Health (Wash). 2024 May 28;2(7):474-485. doi: 10.1021/envhealth.4c00026. eCollection 2024 Jul 19. Environ Health (Wash). 2024. PMID: 39049897 Free PMC article.
-
Analysis of model PM2.5-induced inflammation and cytotoxicity by the combination of a virtual carbon nanoparticle library and computational modeling.Ecotoxicol Environ Saf. 2020 Mar 15;191:110216. doi: 10.1016/j.ecoenv.2020.110216. Epub 2020 Jan 20. Ecotoxicol Environ Saf. 2020. PMID: 31972454 Free PMC article.
-
Advancing Computational Toxicology in the Big Data Era by Artificial Intelligence: Data-Driven and Mechanism-Driven Modeling for Chemical Toxicity.Chem Res Toxicol. 2019 Apr 15;32(4):536-547. doi: 10.1021/acs.chemrestox.8b00393. Epub 2019 Mar 25. Chem Res Toxicol. 2019. PMID: 30907586 Free PMC article. Review.
-
Big Data and Artificial Intelligence Modeling for Drug Discovery.Annu Rev Pharmacol Toxicol. 2020 Jan 6;60:573-589. doi: 10.1146/annurev-pharmtox-010919-023324. Epub 2019 Sep 13. Annu Rev Pharmacol Toxicol. 2020. PMID: 31518513 Free PMC article. Review.
References
-
- Adler S, Basketter D, Creton S, Pelkonen O, van Benthem J, Zuang V, et al. Alternative (non-animal) methods for cosmetics testing: current status and future prospects—2010. Arch Toxicol. 2011;85:367–485. - PubMed
-
- Allen TEH, Goodman JM, Gutsell S, Russell PJ. Defining molecular initiating events in the adverse outcome pathway framework for risk assessment. Chem Res Toxicol. 2014;27:2100–2112. - PubMed
-
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, et al. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29:730–741. - PubMed
-
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

