Two Dimensional Yau-Hausdorff Distance with Applications on Comparison of DNA and Protein Sequences
- PMID: 26384293
- PMCID: PMC4575136
- DOI: 10.1371/journal.pone.0136577
Two Dimensional Yau-Hausdorff Distance with Applications on Comparison of DNA and Protein Sequences
Abstract
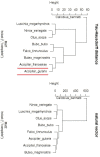
Comparing DNA or protein sequences plays an important role in the functional analysis of genomes. Despite many methods available for sequences comparison, few methods retain the information content of sequences. We propose a new approach, the Yau-Hausdorff method, which considers all translations and rotations when seeking the best match of graphical curves of DNA or protein sequences. The complexity of this method is lower than that of any other two dimensional minimum Hausdorff algorithm. The Yau-Hausdorff method can be used for measuring the similarity of DNA sequences based on two important tools: the Yau-Hausdorff distance and graphical representation of DNA sequences. The graphical representations of DNA sequences conserve all sequence information and the Yau-Hausdorff distance is mathematically proved as a true metric. Therefore, the proposed distance can preciously measure the similarity of DNA sequences. The phylogenetic analyses of DNA sequences by the Yau-Hausdorff distance show the accuracy and stability of our approach in similarity comparison of DNA or protein sequences. This study demonstrates that Yau-Hausdorff distance is a natural metric for DNA and protein sequences with high level of stability. The approach can be also applied to similarity analysis of protein sequences by graphic representations, as well as general two dimensional shape matching.
Conflict of interest statement
Figures









References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215(3):403–410. - PubMed
-
- Zou Q, Hu Q, Guo M, Wang G. HAlign: Fast multiple similar DNA/RNA sequence alignment based on the centre star strategy. Bioinformatics. 2015;p. btv177. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

