A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA
- PMID: 26384420
- PMCID: PMC4737177
- DOI: 10.1093/nar/gkv903
A versatile toolbox for posttranscriptional chemical labeling and imaging of RNA
Abstract
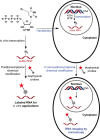
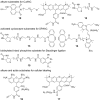
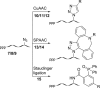
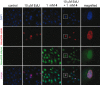
Cellular RNA labeling strategies based on bioorthogonal chemical reactions are much less developed in comparison to glycan, protein and DNA due to its inherent instability and lack of effective methods to introduce bioorthogonal reactive functionalities (e.g. azide) into RNA. Here we report the development of a simple and modular posttranscriptional chemical labeling and imaging technique for RNA by using a novel toolbox comprised of azide-modified UTP analogs. These analogs facilitate the enzymatic incorporation of azide groups into RNA, which can be posttranscriptionally labeled with a variety of probes by click and Staudinger reactions. Importantly, we show for the first time the specific incorporation of azide groups into cellular RNA by endogenous RNA polymerases, which enabled the imaging of newly transcribing RNA in fixed and in live cells by click reactions. This labeling method is practical and provides a new platform to study RNA in vitro and in cells.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
Posttranscriptional chemical functionalization of azide-modified oligoribonucleotides by bioorthogonal click and Staudinger reactions.Chem Commun (Camb). 2012 Jan 14;48(4):498-500. doi: 10.1039/c1cc15659d. Epub 2011 Oct 17. Chem Commun (Camb). 2012. PMID: 22006199
-
Imaging Newly Transcribed RNA in Cells by Using a Clickable Azide-Modified UTP Analog.Methods Mol Biol. 2018;1649:359-371. doi: 10.1007/978-1-4939-7213-5_24. Methods Mol Biol. 2018. PMID: 29130210
-
A clickable UTP analog for the posttranscriptional chemical labeling and imaging of RNA.Org Biomol Chem. 2016 Jun 28;14(24):5832-42. doi: 10.1039/c6ob00576d. Epub 2016 May 13. Org Biomol Chem. 2016. PMID: 27173127
-
Posttranscriptional chemical labeling of RNA by using bioorthogonal chemistry.Methods. 2017 May 1;120:28-38. doi: 10.1016/j.ymeth.2017.02.004. Epub 2017 Feb 17. Methods. 2017. PMID: 28215631 Review.
-
Current approaches for RNA labeling in vitro and in cells based on click reactions.Chembiochem. 2014 Nov 3;15(16):2342-7. doi: 10.1002/cbic.201402240. Epub 2014 Sep 15. Chembiochem. 2014. PMID: 25224574 Review.
Cited by
-
Vinyluridine as a Versatile Chemoselective Handle for the Post-transcriptional Chemical Functionalization of RNA.Bioconjug Chem. 2017 May 17;28(5):1529-1536. doi: 10.1021/acs.bioconjchem.7b00169. Epub 2017 Apr 26. Bioconjug Chem. 2017. PMID: 28406614 Free PMC article.
-
Covalent labeling of nucleic acids.Chem Soc Rev. 2020 Dec 7;49(23):8749-8773. doi: 10.1039/d0cs00600a. Epub 2020 Oct 21. Chem Soc Rev. 2020. PMID: 33084688 Free PMC article. Review.
-
Profiling of the ADP-Ribosylome in Living Cells.Angew Chem Int Ed Engl. 2022 Apr 25;61(18):e202200977. doi: 10.1002/anie.202200977. Epub 2022 Mar 9. Angew Chem Int Ed Engl. 2022. PMID: 35188710 Free PMC article.
-
Comparative Study of Novel Fluorescent Cyanine Nucleotides: Hybridization Analysis of Labeled PCR Products Using a Biochip.J Fluoresc. 2017 Nov;27(6):2001-2016. doi: 10.1007/s10895-017-2139-6. Epub 2017 Jul 28. J Fluoresc. 2017. PMID: 28752470
-
Visualization of newly synthesized neuronal RNA in vitro and in vivo using click-chemistry.RNA Biol. 2017 Jan 2;14(1):20-28. doi: 10.1080/15476286.2016.1251541. Epub 2016 Nov 1. RNA Biol. 2017. PMID: 27801616 Free PMC article.
References
-
- Wachowius F., Höbartner C. Chemical RNA modifications for studies of RNA structure and dynamics. Chembiochem. 2010;11:469–480. - PubMed
-
- Baker M. RNA imaging in situ. Nat. Methods. 2012;9:787–790.
-
- Haukenes G., Szilvay A.-M., Brokstad K.A., Kalland K.-H. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using a liposome transfection reagent. Biotechniques. 1997;22:308–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

