De Novo Assembly and Characterization of Four Anthozoan (Phylum Cnidaria) Transcriptomes
- PMID: 26384772
- PMCID: PMC4632063
- DOI: 10.1534/g3.115.020164
De Novo Assembly and Characterization of Four Anthozoan (Phylum Cnidaria) Transcriptomes
Abstract
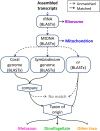
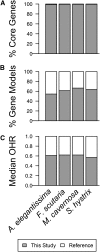
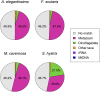
Many nonmodel species exemplify important biological questions but lack the sequence resources required to study the genes and genomic regions underlying traits of interest. Reef-building corals are famously sensitive to rising seawater temperatures, motivating ongoing research into their stress responses and long-term prospects in a changing climate. A comprehensive understanding of these processes will require extending beyond the sequenced coral genome (Acropora digitifera) to encompass diverse coral species and related anthozoans. Toward that end, we have assembled and annotated reference transcriptomes to develop catalogs of gene sequences for three scleractinian corals (Fungia scutaria, Montastraea cavernosa, Seriatopora hystrix) and a temperate anemone (Anthopleura elegantissima). High-throughput sequencing of cDNA libraries produced ~20-30 million reads per sample, and de novo assembly of these reads produced ~75,000-110,000 transcripts from each sample with size distributions (mean ~1.4 kb, N50 ~2 kb), comparable to the distribution of gene models from the coral genome (mean ~1.7 kb, N50 ~2.2 kb). Each assembly includes matches for more than half the gene models from A. digitifera (54-67%) and many reasonably complete transcripts (~5300-6700) spanning nearly the entire gene (ortholog hit ratios ≥0.75). The catalogs of gene sequences developed in this study made it possible to identify hundreds to thousands of orthologs across diverse scleractinian species and related taxa. We used these sequences for phylogenetic inference, recovering known relationships and demonstrating superior performance over phylogenetic trees constructed using single mitochondrial loci. The resources developed in this study provide gene sequences and genetic markers for several anthozoan species. To enhance the utility of these resources for the research community, we developed searchable databases enabling researchers to rapidly recover sequences for genes of interest. Our analysis of de novo assembly quality highlights metrics that we expect will be useful for evaluating the relative quality of other de novo transcriptome assemblies. The identification of orthologous sequences and phylogenetic reconstruction demonstrates the feasibility of these methods for clarifying the substantial uncertainties in the existing scleractinian phylogeny.
Keywords: coral; database; nonmodel system; phylogenomics.
Copyright © 2015 Kitchen et al.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
