Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants
- PMID: 26384972
- PMCID: PMC4575454
- DOI: 10.1186/s12870-015-0596-0
Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants
Abstract
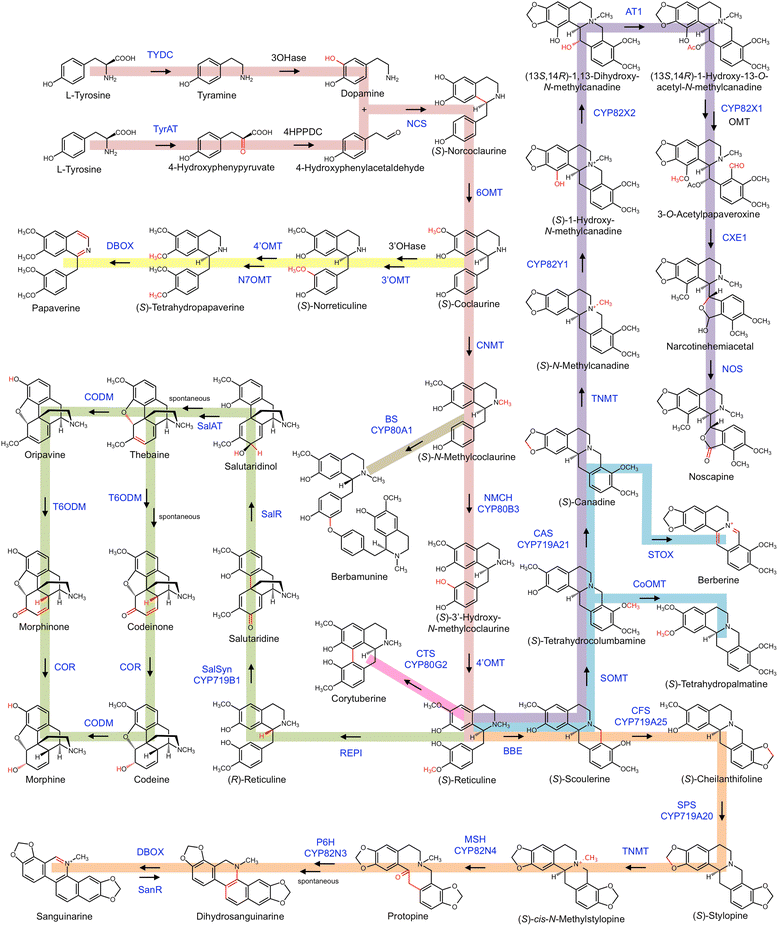
Background: Benzylisoquinoline alkaloids (BIAs) represent a diverse class of plant specialized metabolites sharing a common biosynthetic origin beginning with tyrosine. Many BIAs have potent pharmacological activities, and plants accumulating them boast long histories of use in traditional medicine and cultural practices. The decades-long focus on a select number of plant species as model systems has allowed near or full elucidation of major BIA pathways, including those of morphine, sanguinarine and berberine. However, this focus has created a dearth of knowledge surrounding non-model species, which also are known to accumulate a wide-range of BIAs but whose biosynthesis is thus far entirely unexplored. Further, these non-model species represent a rich source of catalyst diversity valuable to plant biochemists and emerging synthetic biology efforts.
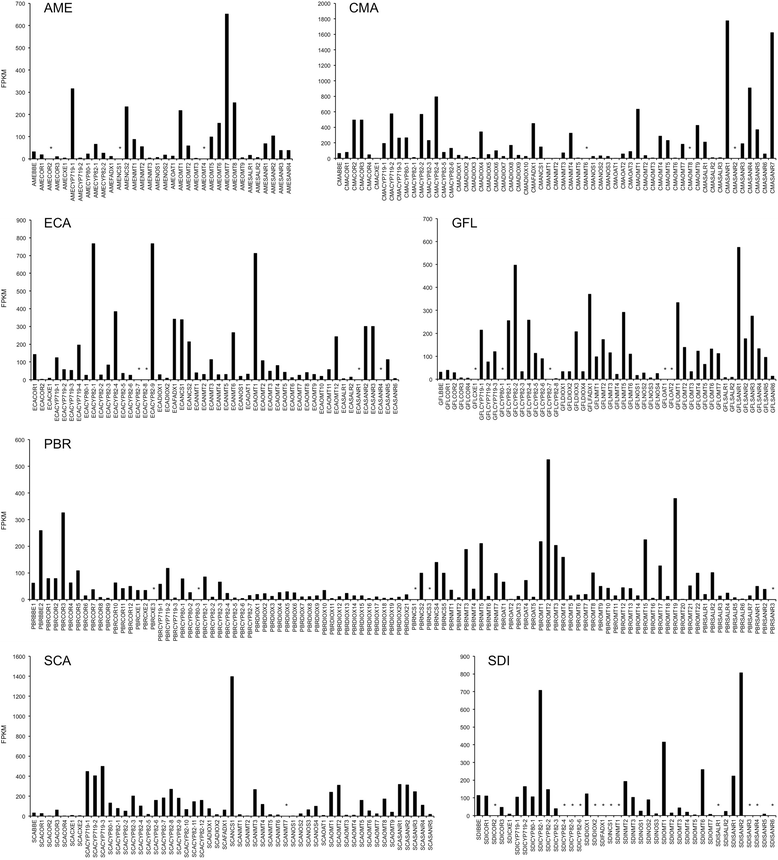
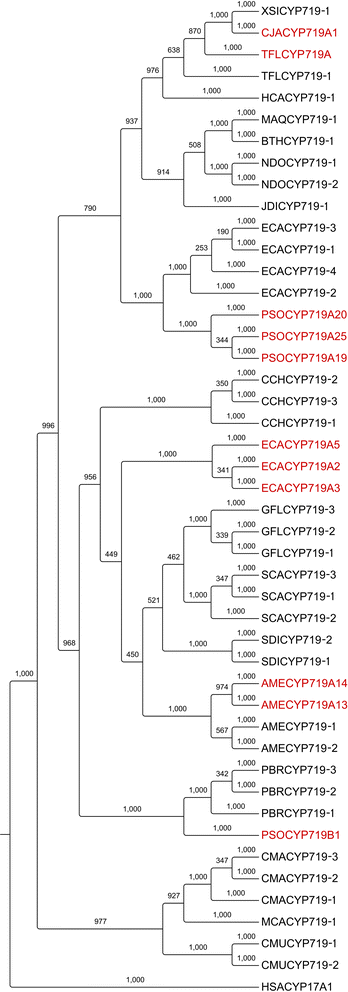
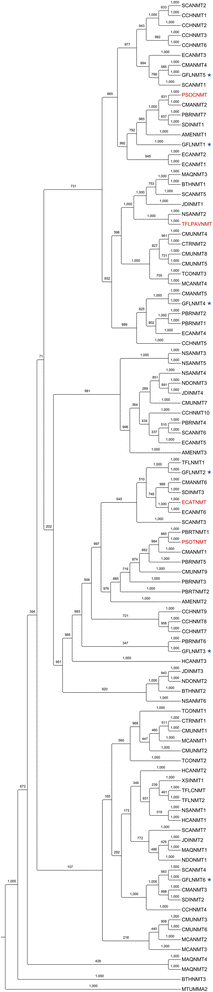
Results: In order to access the genetic diversity of non-model plants accumulating BIAs, we selected 20 species representing 4 families within the Ranunculales. RNA extracted from each species was processed for analysis by both 1) Roche GS-FLX Titanium and 2) Illumina GA/HiSeq platforms, generating a total of 40 deep-sequencing transcriptome libraries. De novo assembly, annotation and subsequent full-length coding sequence (CDS) predictions indicated greater success for most species using the Illumina-based platform. Assembled data for each transcriptome were deposited into an established web-based BLAST portal ( www.phytometasyn.ca) to allow public access. Homology-based mining of libraries using BIA-biosynthetic enzymes as queries yielded ~850 gene candidates potentially involved in alkaloid biosynthesis. Expression analysis of these candidates was performed using inter-library FPKM normalization methods. These expression data provide a basis for the rational selection of gene candidates, and suggest possible metabolic bottlenecks within BIA metabolism. Phylogenetic analysis was performed for each of 15 different enzyme/protein groupings, highlighting many novel genes with potential involvement in the formation of one or more alkaloid types, including morphinan, aporphine, and phthalideisoquinoline alkaloids. Transcriptome resources were used to design and execute a case study of candidate N-methyltransferases (NMTs) from Glaucium flavum, which revealed predicted and novel enzyme activities.
Conclusions: This study establishes an essential resource for the isolation and discovery of 1) functional homologues and 2) entirely novel catalysts within BIA metabolism. Functional analysis of G. flavum NMTs demonstrated the utility of this resource and underscored the importance of empirical determination of proposed enzymatic function. Publically accessible, fully annotated, BLAST-accessible transcriptomes were not previously available for most species included in this report, despite the rich repertoire of bioactive alkaloids found in these plants and their importance to traditional medicine. The results presented herein provide essential sequence information and inform experimental design for the continued elucidation of BIA metabolism.
Figures
References
-
- Beaudoin GAW, Facchini PJ. Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem Biophys Res Commum. 2013;431:597–603. - PubMed
-
- Blaskó G, Gula DJ, Shamma M. The phthalideisoquinoline alkaloids. J Nat Prod. 1982;45:105–122. doi: 10.1021/np50020a001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

