Fanconi anemia cells with unrepaired DNA damage activate components of the checkpoint recovery process
- PMID: 26385365
- PMCID: PMC4575447
- DOI: 10.1186/s12976-015-0011-4
Fanconi anemia cells with unrepaired DNA damage activate components of the checkpoint recovery process
Abstract
Background: The FA/BRCA pathway repairs DNA interstrand crosslinks. Mutations in this pathway cause Fanconi anemia (FA), a chromosome instability syndrome with bone marrow failure and cancer predisposition. Upon DNA damage, normal and FA cells inhibit the cell cycle progression, until the G2/M checkpoint is turned off by the checkpoint recovery, which becomes activated when the DNA damage has been repaired. Interestingly, highly damaged FA cells seem to override the G2/M checkpoint. In this study we explored with a Boolean network model and key experiments whether checkpoint recovery activation occurs in FA cells with extensive unrepaired DNA damage.
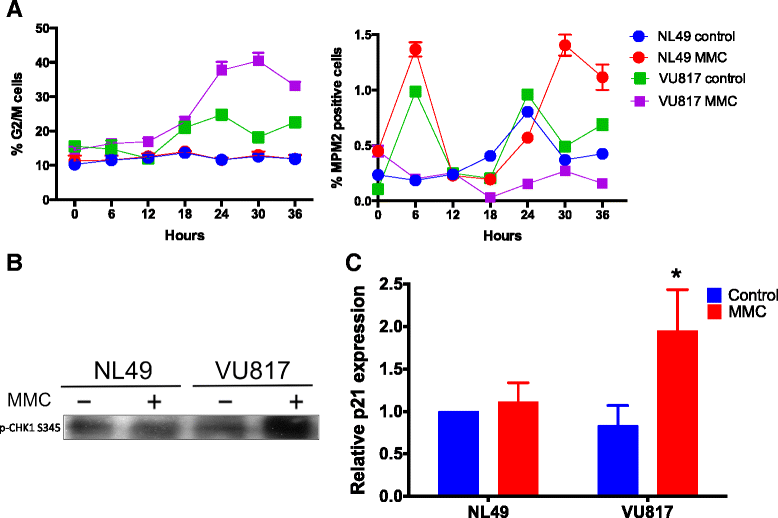
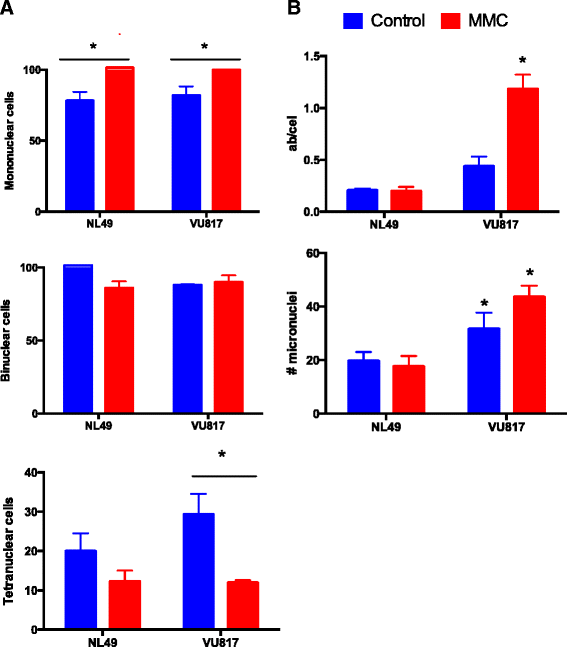
Methods: We performed synchronous/asynchronous simulations of the FA/BRCA pathway Boolean network model. FA-A and normal lymphoblastoid cell lines were used to study checkpoint and checkpoint recovery activation after DNA damage induction. The experimental approach included flow cytometry cell cycle analysis, cell division tracking, chromosome aberration analysis and gene expression analysis through qRT-PCR and western blot.
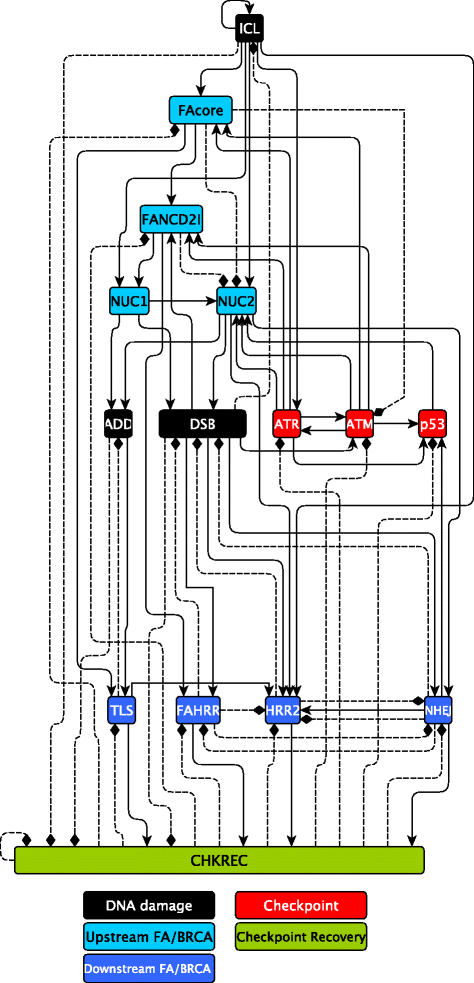
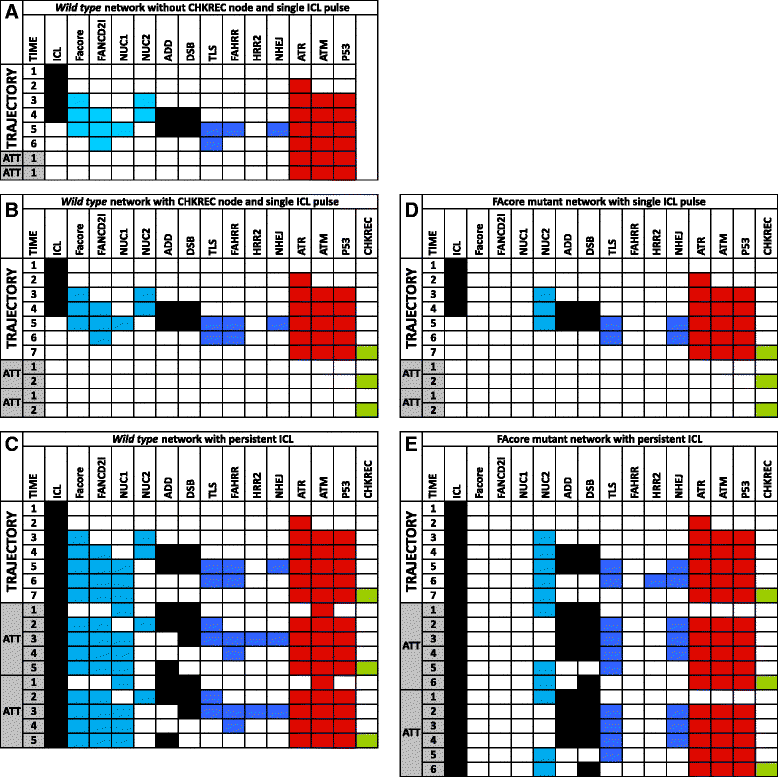
Results: Computational simulations suggested that in FA mutants checkpoint recovery activity inhibits the checkpoint components despite unrepaired DNA damage, a behavior that we did not observed in wild-type simulations. This result implies that FA cells would eventually reenter the cell cycle after a DNA damage induced G2/M checkpoint arrest, but before the damage has been fixed. We observed that FA-A cells activate the G2/M checkpoint and arrest in G2 phase, but eventually reach mitosis and divide with unrepaired DNA damage, thus resolving the initial checkpoint arrest. Based on our model result we look for ectopic activity of checkpoint recovery components. We found that checkpoint recovery components, such as PLK1, are expressed to a similar extent as normal undamaged cells do, even though FA-A cells harbor highly damaged DNA.
Conclusions: Our results show that FA cells, despite extensive DNA damage, do not loss the capacity to express the transcriptional and protein components of checkpoint recovery that might eventually allow their division with unrepaired DNA damage. This might allow cell survival but increases the genomic instability inherent to FA individuals and promotes cancer.
Figures




Similar articles
-
WIP1 Contributes to the Adaptation of Fanconi Anemia Cells to DNA Damage as Determined by the Regulatory Network of the Fanconi Anemia and Checkpoint Recovery Pathways.Front Genet. 2019 May 3;10:411. doi: 10.3389/fgene.2019.00411. eCollection 2019. Front Genet. 2019. PMID: 31130988 Free PMC article.
-
Differential expression of TP53 associated genes in Fanconi anemia cells after mitomycin C and hydroxyurea treatment.Mutat Res. 2008 Oct 30;656(1-2):1-7. doi: 10.1016/j.mrgentox.2008.06.012. Epub 2008 Jul 5. Mutat Res. 2008. PMID: 18647660
-
A role for the Fanconi anemia C protein in maintaining the DNA damage-induced G2 checkpoint.J Biol Chem. 2004 Dec 3;279(49):50986-93. doi: 10.1074/jbc.M407160200. Epub 2004 Sep 17. J Biol Chem. 2004. PMID: 15377654
-
Alpha-fetoprotein and Fanconi Anemia: Relevance to DNA Repair and Breast Cancer Susceptibility.Fetal Pediatr Pathol. 2017 Feb;36(1):49-61. doi: 10.1080/15513815.2016.1225873. Epub 2016 Oct 3. Fetal Pediatr Pathol. 2017. PMID: 27690720 Review.
-
[Pathogenesis of Fanconi anemia: FA-BRCA network -- review].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2009 Jun;17(3):805-9. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2009. PMID: 19549413 Review. Chinese.
Cited by
-
Multiomic Analysis of Cereblon Expression and Its Prognostic Value in Kidney Renal Clear Cell Carcinoma, Lung Adenocarcinoma, and Skin Cutaneous Melanoma.J Pers Med. 2021 Apr 1;11(4):263. doi: 10.3390/jpm11040263. J Pers Med. 2021. PMID: 33916291 Free PMC article.
-
Antifragility Predicts the Robustness and Evolvability of Biological Networks through Multi-Class Classification with a Convolutional Neural Network.Entropy (Basel). 2020 Sep 4;22(9):986. doi: 10.3390/e22090986. Entropy (Basel). 2020. PMID: 33286756 Free PMC article.
-
DNA Damage as a Driver for Growth Delay: Chromosome Instability Syndromes with Intrauterine Growth Retardation.Biomed Res Int. 2017;2017:8193892. doi: 10.1155/2017/8193892. Epub 2017 Nov 12. Biomed Res Int. 2017. PMID: 29238724 Free PMC article. Review.
-
Boolean network modeling in systems pharmacology.J Pharmacokinet Pharmacodyn. 2018 Feb;45(1):159-180. doi: 10.1007/s10928-017-9567-4. Epub 2018 Jan 6. J Pharmacokinet Pharmacodyn. 2018. PMID: 29307099 Free PMC article. Review.
-
Chromosome Instability in Fanconi Anemia: From Breaks to Phenotypic Consequences.Genes (Basel). 2020 Dec 21;11(12):1528. doi: 10.3390/genes11121528. Genes (Basel). 2020. PMID: 33371494 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

