Molecular mechanisms of ribosomal protein gene coregulation
- PMID: 26385964
- PMCID: PMC4579351
- DOI: 10.1101/gad.268896.115
Molecular mechanisms of ribosomal protein gene coregulation
Abstract
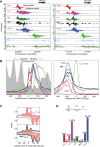
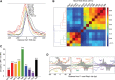
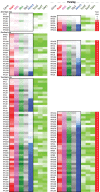
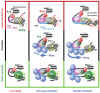
The 137 ribosomal protein genes (RPGs) of Saccharomyces provide a model for gene coregulation. We examined the positional and functional organization of their regulators (Rap1 [repressor activator protein 1], Fhl1, Ifh1, Sfp1, and Hmo1), the transcription machinery (TFIIB, TFIID, and RNA polymerase II), and chromatin at near-base-pair resolution using ChIP-exo, as RPGs are coordinately reprogrammed. Where Hmo1 is enriched, Fhl1, Ifh1, Sfp1, and Hmo1 cross-linked broadly to promoter DNA in an RPG-specific manner and demarcated by general minor groove widening. Importantly, Hmo1 extended 20-50 base pairs (bp) downstream from Fhl1. Upon RPG repression, Fhl1 remained in place. Hmo1 dissociated, which was coupled to an upstream shift of the +1 nucleosome, as reflected by the Hmo1 extension and core promoter region. Fhl1 and Hmo1 may create two regulatable and positionally distinct barriers, against which chromatin remodelers position the +1 nucleosome into either an activating or a repressive state. Consistent with in vitro studies, we found that specific TFIID subunits, in addition to cross-linking at the core promoter, made precise cross-links at Rap1 sites, which we interpret to reflect native Rap1-TFIID interactions. Our findings suggest how sequence-specific DNA binding regulates nucleosome positioning and transcription complex assembly >300 bp away and how coregulation coevolved with coding sequences.
Keywords: ChIP-seq; chromatin; gene regulation mechanisms; transcription factor.
© 2015 Reja et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription.Genes Dev. 2014 Aug 1;28(15):1695-709. doi: 10.1101/gad.244434.114. Genes Dev. 2014. PMID: 25085421 Free PMC article.
-
Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Oct;27(19):6686-705. doi: 10.1128/MCB.00876-07. Epub 2007 Jul 23. Mol Cell Biol. 2007. PMID: 17646381 Free PMC article.
-
An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae.Mol Cell Biol. 2006 May;26(9):3672-9. doi: 10.1128/MCB.26.9.3672-3679.2006. Mol Cell Biol. 2006. PMID: 16612005 Free PMC article.
-
Transcriptional control of ribosome biogenesis in yeast: links to growth and stress signals.Biochem Soc Trans. 2021 Aug 27;49(4):1589-1599. doi: 10.1042/BST20201136. Biochem Soc Trans. 2021. PMID: 34240738 Free PMC article. Review.
-
Physiological function of FKBP12, a primary target of rapamycin/FK506: a newly identified role in transcription of ribosomal protein genes in yeast.Curr Genet. 2021 Jun;67(3):383-388. doi: 10.1007/s00294-020-01142-3. Epub 2021 Jan 12. Curr Genet. 2021. PMID: 33438053 Review.
Cited by
-
Hmo1: A versatile member of the high mobility group box family of chromosomal architecture proteins.World J Biol Chem. 2024 Aug 12;15(1):97938. doi: 10.4331/wjbc.v15.i1.97938. World J Biol Chem. 2024. PMID: 39156122 Free PMC article. Review.
-
Differential requirements for Gcn5 and NuA4 HAT activities in the starvation-induced versus basal transcriptomes.Nucleic Acids Res. 2023 May 8;51(8):3696-3721. doi: 10.1093/nar/gkad099. Nucleic Acids Res. 2023. PMID: 36864781 Free PMC article.
-
Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters.EMBO J. 2017 Feb 1;36(3):274-290. doi: 10.15252/embj.201695621. Epub 2016 Dec 15. EMBO J. 2017. PMID: 27979920 Free PMC article.
-
Identification of a transcriptional activation domain in yeast repressor activator protein 1 (Rap1) using an altered DNA-binding specificity variant.J Biol Chem. 2017 Apr 7;292(14):5705-5723. doi: 10.1074/jbc.M117.779181. Epub 2017 Feb 14. J Biol Chem. 2017. PMID: 28196871 Free PMC article.
-
Regulation of ribosomal protein genes: An ordered anarchy.Wiley Interdiscip Rev RNA. 2021 May;12(3):e1632. doi: 10.1002/wrna.1632. Epub 2020 Oct 10. Wiley Interdiscip Rev RNA. 2021. PMID: 33038057 Free PMC article. Review.
References
-
- Cherel I, Thuriaux P. 1995. The IFH1 gene product interacts with a fork head protein in Saccharomyces cerevisiae. Yeast 11: 261–270. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
