Iron response regulator protein IrrB in Magnetospirillum gryphiswaldense MSR-1 helps control the iron/oxygen balance, oxidative stress tolerance, and magnetosome formation
- PMID: 26386052
- PMCID: PMC4651088
- DOI: 10.1128/AEM.02585-15
Iron response regulator protein IrrB in Magnetospirillum gryphiswaldense MSR-1 helps control the iron/oxygen balance, oxidative stress tolerance, and magnetosome formation
Abstract
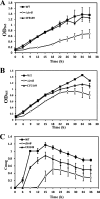
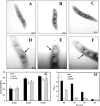
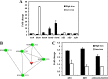
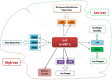
Magnetotactic bacteria are capable of forming nanosized, membrane-enclosed magnetosomes under iron-rich and oxygen-limited conditions. The complete genomic sequence of Magnetospirillum gryphiswaldense strain MSR-1 has been analyzed and found to contain five fur homologue genes whose protein products are predicted to be involved in iron homeostasis and the response to oxidative stress. Of these, only the MGMSRv2_3149 gene (irrB) was significantly downregulated under high-iron and low-oxygen conditions, during the transition of cell growth from the logarithmic to the stationary phase. The encoded protein, IrrB, containing the conserved HHH motif, was identified as an iron response regulator (Irr) protein belonging to the Fur superfamily. To investigate the function of IrrB, we constructed an irrB deletion mutant (ΔirrB). The levels of cell growth and magnetosome formation were lower in the ΔirrB strain than in the wild type (WT) under both high-iron and low-iron conditions. The ΔirrB strain also showed lower levels of iron uptake and H2O2 tolerance than the WT. Quantitative real-time reverse transcription-PCR analysis indicated that the irrB mutation reduced the expression of numerous genes involved in iron transport, iron storage, heme biosynthesis, and Fe-S cluster assembly. Transcription studies of the other fur homologue genes in the ΔirrB strain indicated complementary functions of the Fur proteins in MSR-1. IrrB appears to be directly responsible for iron metabolism and homeostasis and to be indirectly involved in magnetosome formation. We propose two IrrB-regulated networks (under high- and low-iron conditions) in MSR-1 cells that control the balance of iron and oxygen metabolism and account for the coexistence of five Fur homologues.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Physiological characteristics of Magnetospirillum gryphiswaldense MSR-1 that control cell growth under high-iron and low-oxygen conditions.Sci Rep. 2017 Jun 5;7(1):2800. doi: 10.1038/s41598-017-03012-4. Sci Rep. 2017. PMID: 28584275 Free PMC article.
-
Fur in Magnetospirillum gryphiswaldense influences magnetosomes formation and directly regulates the genes involved in iron and oxygen metabolism.PLoS One. 2012;7(1):e29572. doi: 10.1371/journal.pone.0029572. Epub 2012 Jan 4. PLoS One. 2012. PMID: 22238623 Free PMC article.
-
Expression patterns of key iron and oxygen metabolism genes during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1.FEMS Microbiol Lett. 2013 Oct;347(2):163-72. doi: 10.1111/1574-6968.12234. Epub 2013 Sep 6. FEMS Microbiol Lett. 2013. PMID: 23937222
-
Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense.Arch Microbiol. 2004 Jan;181(1):1-7. doi: 10.1007/s00203-003-0631-7. Epub 2003 Dec 11. Arch Microbiol. 2004. PMID: 14668979 Review.
-
Bacterial magnetosome and its potential application.Microbiol Res. 2017 Oct;203:19-28. doi: 10.1016/j.micres.2017.06.005. Epub 2017 Jun 20. Microbiol Res. 2017. PMID: 28754204 Review.
Cited by
-
The FUR-like regulators PerRA and PerRB integrate a complex regulatory network that promotes mammalian host-adaptation and virulence of Leptospira interrogans.PLoS Pathog. 2021 Dec 2;17(12):e1009078. doi: 10.1371/journal.ppat.1009078. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34855918 Free PMC article.
-
Magnetic genes: Studying the genetics of biomineralization in magnetotactic bacteria.PLoS Genet. 2020 Feb 13;16(2):e1008499. doi: 10.1371/journal.pgen.1008499. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32053597 Free PMC article. Review.
-
Nitric oxide sensor NsrR is the key direct regulator of magnetosome formation and nitrogen metabolism in Magnetospirillum.Nucleic Acids Res. 2024 Apr 12;52(6):2924-2941. doi: 10.1093/nar/gkad1230. Nucleic Acids Res. 2024. PMID: 38197240 Free PMC article.
-
Physiological characteristics of Magnetospirillum gryphiswaldense MSR-1 that control cell growth under high-iron and low-oxygen conditions.Sci Rep. 2017 Jun 5;7(1):2800. doi: 10.1038/s41598-017-03012-4. Sci Rep. 2017. PMID: 28584275 Free PMC article.
-
Magnetosome biogenesis in magnetotactic bacteria.Nat Rev Microbiol. 2016 Sep 13;14(10):621-37. doi: 10.1038/nrmicro.2016.99. Nat Rev Microbiol. 2016. PMID: 27620945 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

