Contribution of the distal pocket residue to the acyl-chain-length specificity of (R)-specific enoyl-coenzyme A hydratases from Pseudomonas spp
- PMID: 26386053
- PMCID: PMC4651097
- DOI: 10.1128/AEM.02412-15
Contribution of the distal pocket residue to the acyl-chain-length specificity of (R)-specific enoyl-coenzyme A hydratases from Pseudomonas spp
Abstract
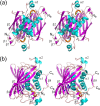
(R)-Specific enoyl-coenzyme A (enoyl-CoA) hydratases (PhaJs) are capable of supplying monomers from fatty acid β-oxidation to polyhydroxyalkanoate (PHA) biosynthesis. PhaJ1Pp from Pseudomonas putida showed broader substrate specificity than did PhaJ1Pa from Pseudomonas aeruginosa, despite sharing 67% amino acid sequence identity. In this study, the substrate specificity characteristics of two Pseudomonas PhaJ1 enzymes were investigated by site-directed mutagenesis, chimeragenesis, X-ray crystallographic analysis, and homology modeling. In PhaJ1Pp, the replacement of valine with isoleucine at position 72 resulted in an increased preference for enoyl-coenzyme A (CoA) elements with shorter chain lengths. Conversely, at the same position in PhaJ1Pa, the replacement of isoleucine with valine resulted in an increased preference for enoyl-CoAs with longer chain lengths. These changes suggest a narrowing and broadening in the substrate specificity range of the PhaJ1Pp and PhaJ1Pa mutants, respectively. However, the substrate specificity remains broader in PhaJ1Pp than in PhaJ1Pa. Additionally, three chimeric PhaJ1 enzymes, composed from PhaJ1Pp and PhaJ1Pa, all showed significant hydratase activity, and their substrate preferences were within the range exhibited by the parental PhaJ1 enzymes. The crystal structure of PhaJ1Pa was determined at a resolution of 1.7 Å, and subsequent homology modeling of PhaJ1Pp revealed that in the acyl-chain binding pocket, the amino acid at position 72 was the only difference between the two structures. These results indicate that the chain-length specificity of PhaJ1 is determined mainly by the bulkiness of the amino acid residue at position 72, but that other factors, such as structural fluctuations, also affect specificity.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Sudesh K, Abe H, Doi Y. 2000. Synthesis, structure and properties of polyhydroxyalkanoate: biological polyesters. Prog Polym Sci 25:1503–1555. doi:10.1016/S0079-6700(00)00035-6. - DOI
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

