Genomic evidence reveals numerous Salmonella enterica serovar Newport reintroduction events in Suwannee watershed irrigation ponds
- PMID: 26386063
- PMCID: PMC4644655
- DOI: 10.1128/AEM.02179-15
Genomic evidence reveals numerous Salmonella enterica serovar Newport reintroduction events in Suwannee watershed irrigation ponds
Abstract
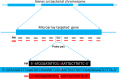
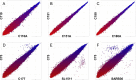
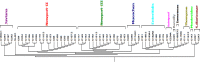
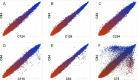
Our previous work indicated a predominance (56.8%) of Salmonella enterica serovar Newport among isolates recovered from irrigation ponds used in produce farms over a 2-year period (B. Li et al., Appl Environ Microbiol 80:6355-6365, http://dx.doi.org/10.1128/AEM.02063-14). This observation provided a valuable set of metrics to explore an underaddressed issue of environmental survival of Salmonella by DNA microarray. Microarray analysis correctly identified all the isolates (n = 53) and differentiated the S. Newport isolates into two phylogenetic lineages (S. Newport II and S. Newport III). Serovar distribution analysis showed no instances where the same serovar was recovered from a pond for more than a month. Furthermore, during the study, numerous isolates with an indistinguishable genotype were recovered from different ponds as far as 180 km apart for time intervals as long as 2 years. Although isolates within either lineage were phylogenetically related as determined by microarray analysis, subtle genotypic differences were detected within the lineages, suggesting that isolates in either lineage could have come from several unique hosts. For example, strains in four different subgroups (A, B, C, and D) possessed an indistinguishable genotype within their subgroups as measured by gene differences, suggesting that strains in each subgroup shared a common host. Based on this comparative genomic evidence and the spatial and temporal factors, we speculated that the presence of Salmonella in the ponds was likely due to numerous punctuated reintroduction events associated with several different but common hosts in the environment. These findings may have implications for the development of strategies for efficient and safe irrigation to minimize the risk of Salmonella outbreaks associated with fresh produce.
Copyright © 2015 Li et al.
Figures
References
-
- CDC. 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb Mortal Wkly Rep 59:418–422. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

