Global transcriptome and mutagenic analyses of the acid tolerance response of Salmonella enterica serovar Typhimurium
- PMID: 26386064
- PMCID: PMC4651094
- DOI: 10.1128/AEM.02172-15
Global transcriptome and mutagenic analyses of the acid tolerance response of Salmonella enterica serovar Typhimurium
Abstract
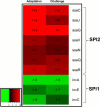
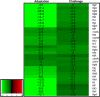
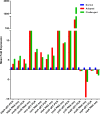
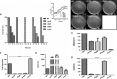
Salmonella enterica serovar Typhimurium (S. Typhimurium) is one of the leading causative agents of food-borne bacterial gastroenteritis. Swift invasion through the intestinal tract and successful establishment in systemic organs are associated with the adaptability of S. Typhimurium to different stress environments. Low-pH stress serves as one of the first lines of defense in mammalian hosts, which S. Typhimurium must efficiently overcome to establish an infection. Therefore, a better understanding of the molecular mechanisms underlying the adaptability of S. Typhimurium to acid stress is highly relevant. In this study, we have performed a transcriptome analysis of S. Typhimurium under the acid tolerance response (ATR) and found a large number of genes (∼47%) to be differentially expressed (more than 1.5-fold or less than -1.5-fold; P < 0.01). Functional annotation revealed differentially expressed genes to be associated with regulation, metabolism, transport and binding, pathogenesis, and motility. Additionally, our knockout analysis of a subset of differentially regulated genes facilitated the identification of proteins that contribute to S. Typhimurium ATR and virulence. Mutants lacking genes encoding the K(+) binding and transport protein KdpA, hypothetical protein YciG, the flagellar hook cap protein FlgD, and the nitrate reductase subunit NarZ were significantly deficient in their ATRs and displayed varied in vitro virulence characteristics. This study offers greater insight into the transcriptome changes of S. Typhimurium under the ATR and provides a framework for further research on the subject.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
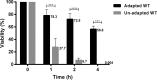

Similar articles
-
The responses of Salmonella enterica serovar Typhimurium to vanillin in apple juice through global transcriptomics.Int J Food Microbiol. 2021 Jun 2;347:109189. doi: 10.1016/j.ijfoodmicro.2021.109189. Epub 2021 Mar 31. Int J Food Microbiol. 2021. PMID: 33838479
-
Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy.Infect Immun. 2013 Jan;81(1):154-65. doi: 10.1128/IAI.01080-12. Epub 2012 Oct 22. Infect Immun. 2013. PMID: 23090959 Free PMC article.
-
FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s).J Bacteriol. 2007 Mar;189(6):2262-73. doi: 10.1128/JB.00726-06. Epub 2007 Jan 12. J Bacteriol. 2007. PMID: 17220229 Free PMC article.
-
Nutritional and metabolic requirements for the infection of HeLa cells by Salmonella enterica serovar Typhimurium.PLoS One. 2014 May 5;9(5):e96266. doi: 10.1371/journal.pone.0096266. eCollection 2014. PLoS One. 2014. PMID: 24797930 Free PMC article.
-
The challenge of relating gene expression to the virulence of Salmonella enterica serovar Typhimurium.Curr Opin Biotechnol. 2011 Apr;22(2):200-10. doi: 10.1016/j.copbio.2011.02.007. Epub 2011 Mar 22. Curr Opin Biotechnol. 2011. PMID: 21388802 Review.
Cited by
-
Metal Sequestration and Antimicrobial Activity of Human Calprotectin Are pH-Dependent.Biochemistry. 2020 Jul 7;59(26):2468-2478. doi: 10.1021/acs.biochem.0c00359. Epub 2020 Jun 22. Biochemistry. 2020. PMID: 32491853 Free PMC article.
-
Survival and adaptative strategies of Enterotoxigenic E. coli (ETEC) to the freshwater environment.Res Sq [Preprint]. 2025 Mar 19:rs.3.rs-6252921. doi: 10.21203/rs.3.rs-6252921/v1. Res Sq. 2025. PMID: 40166005 Free PMC article. Preprint.
-
Beyond Homeostasis: Potassium and Pathogenesis during Bacterial Infections.Infect Immun. 2021 Jun 16;89(7):e0076620. doi: 10.1128/IAI.00766-20. Epub 2021 Jun 16. Infect Immun. 2021. PMID: 33875474 Free PMC article. Review.
-
Global transcriptomic analysis of ethanol tolerance response in Salmonella Enteritidis.Curr Res Food Sci. 2022 May 6;5:798-806. doi: 10.1016/j.crfs.2022.04.011. eCollection 2022. Curr Res Food Sci. 2022. PMID: 35600539 Free PMC article.
-
Genotypic and phenotypic analysis of Salmonella enterica serovar Derby, looking for clues explaining the impairment of egg isolates to cause human disease.Front Microbiol. 2024 Jun 6;15:1357881. doi: 10.3389/fmicb.2024.1357881. eCollection 2024. Front Microbiol. 2024. PMID: 38903793 Free PMC article.
References
-
- Pui CF, Wong WC, Chai LC, Nillian E, Ghazali FM, Cheah YK, Nakaguchi Y, Nishibuchi M, Radu S. 2011. Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control 22:337–342. doi:10.1016/j.foodcont.2010.05.021. - DOI - PMC - PubMed
-
- Álvarez-Ordóñez A, Prieto M, Bernardo A, Hill C, López M. 2012. The acid tolerance response of Salmonella spp.: an adaptive strategy to survive in stressful environments prevailing in foods and the host. Food Res Int 45:482–492. doi:10.1016/j.foodres.2011.04.002. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

