The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells
- PMID: 26387536
- PMCID: PMC6818728
- DOI: 10.1038/onc.2015.325
The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells
Abstract
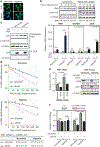
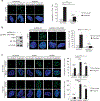
Previously, it has been shown that pancreatic ductal adenocarcinoma (PDA) tumors exhibit high levels of hypoxia, characterized by low oxygen pressure (pO2) and decreased O2 intracellular perfusion. Chronic hypoxia is strongly associated with resistance to cytotoxic chemotherapy and chemoradiation in an understudied phenomenon known as hypoxia-induced chemoresistance. The hypoxia-inducible, pro-oncogenic, serine-threonine kinase PIM1 (Proviral Integration site for Moloney murine leukemia virus 1) has emerged as a key regulator of hypoxia-induced chemoresistance in PDA and other cancers. Although its role in therapeutic resistance has been described previously, the molecular mechanism behind PIM1 overexpression in PDA is unknown. Here, we demonstrate that cis-acting AU-rich elements (ARE) present within a 38-base pair region of the PIM1 mRNA 3'-untranslated region mediate a regulatory interaction with the mRNA stability factor HuR (Hu antigen R) in the context of tumor hypoxia. Predominantly expressed in the nucleus in PDA cells, HuR translocates to the cytoplasm in response to hypoxic stress and stabilizes the PIM1 mRNA transcript, resulting in PIM1 protein overexpression. A reverse-phase protein array revealed that HuR-mediated regulation of PIM1 protects cells from hypoxic stress through phosphorylation and inactivation of the apoptotic effector BAD and activation of MEK1/2. Importantly, pharmacological inhibition of HuR by MS-444 inhibits HuR homodimerization and its cytoplasmic translocation, abrogates hypoxia-induced PIM1 overexpression and markedly enhances PDA cell sensitivity to oxaliplatin and 5-fluorouracil under physiologic low oxygen conditions. Taken together, these results support the notion that HuR has prosurvival properties in PDA cells by enabling them with growth advantages in stressful tumor microenvironment niches. Accordingly, these studies provide evidence that therapeutic disruption of HuR's regulation of PIM1 may be a key strategy in breaking an elusive chemotherapeutic resistance mechanism acquired by PDA cells that reside in hypoxic PDA microenvironments.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures





Similar articles
-
HuR Contributes to TRAIL Resistance by Restricting Death Receptor 4 Expression in Pancreatic Cancer Cells.Mol Cancer Res. 2016 Jul;14(7):599-611. doi: 10.1158/1541-7786.MCR-15-0448. Epub 2016 Apr 6. Mol Cancer Res. 2016. PMID: 27053682 Free PMC article.
-
HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells.Cancer Res. 2014 Feb 15;74(4):1128-40. doi: 10.1158/0008-5472.CAN-13-1915. Cancer Res. 2014. PMID: 24536047 Free PMC article.
-
Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells.Oncotarget. 2015 Sep 29;6(29):27312-31. doi: 10.18632/oncotarget.4743. Oncotarget. 2015. PMID: 26314962 Free PMC article.
-
Targeting the RNA-Binding Protein HuR in Cancer.Cancer Res. 2023 Nov 1;83(21):3507-3516. doi: 10.1158/0008-5472.CAN-23-0972. Cancer Res. 2023. PMID: 37683260 Review.
-
Inhibition of Caspase-2 Translation by the mRNA Binding Protein HuR: A Novel Path of Therapy Resistance in Colon Carcinoma Cells?Cells. 2019 Jul 30;8(8):797. doi: 10.3390/cells8080797. Cells. 2019. PMID: 31366165 Free PMC article. Review.
Cited by
-
Post-transcriptional regulation of cytokine expression and signaling.Curr Trends Immunol. 2018;19:33-40. Curr Trends Immunol. 2018. PMID: 30568341 Free PMC article.
-
RNA-binding protein HuR inhibition induces multiple programmed cell death in breast and prostate cancer.Cell Commun Signal. 2024 Dec 3;22(1):580. doi: 10.1186/s12964-024-01916-z. Cell Commun Signal. 2024. PMID: 39627778 Free PMC article.
-
AFP shields hepatocellular carcinoma from macrophage phagocytosis by regulating HuR-mediated CD47 translocation in cellular membrane.Transl Oncol. 2025 Feb;52:102240. doi: 10.1016/j.tranon.2024.102240. Epub 2024 Dec 11. Transl Oncol. 2025. PMID: 39667226 Free PMC article.
-
Challenges in Therapeutically Targeting the RNA-Recognition Motif.Wiley Interdiscip Rev RNA. 2024 Nov-Dec;15(6):e1877. doi: 10.1002/wrna.1877. Wiley Interdiscip Rev RNA. 2024. PMID: 39668490 Free PMC article. Review.
-
Hallmarks of cancer and AU-rich elements.Wiley Interdiscip Rev RNA. 2017 Jan;8(1):e1368. doi: 10.1002/wrna.1368. Epub 2016 Jun 1. Wiley Interdiscip Rev RNA. 2017. PMID: 27251431 Free PMC article. Review.
References
-
- ACS. Cancer Facts & Figures 2013. American Cancer Society: Atlanta, GA, USA, 2014. Available at http://www.cancer.org/acs/groups/content/@editorial/documents/document/a....
-
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74: 2913–2921. - PubMed
-
- Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011; 364: 1817–1825. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

