Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells
- PMID: 26387834
- PMCID: PMC4576406
- DOI: 10.1186/s13059-015-0739-5
Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells
Abstract
Background: Hematopoietic stem cells (HSCs) are a rare cell type with the ability of long-term self-renewal and multipotency to reconstitute all blood lineages. HSCs are typically purified from the bone marrow using cell surface markers. Recent studies have identified significant cellular heterogeneities in the HSC compartment with subsets of HSCs displaying lineage bias. We previously discovered that the transcription factor Bcl11a has critical functions in the lymphoid development of the HSC compartment.
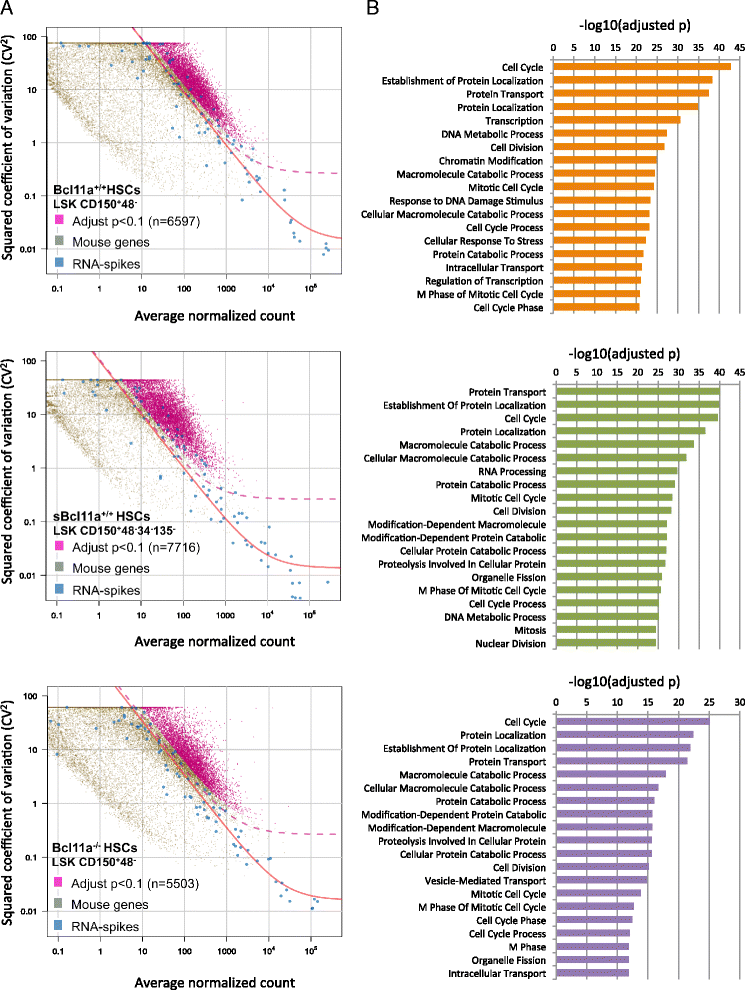
Results: In this report, we employ single-cell transcriptomic analysis to dissect the molecular heterogeneities in HSCs. We profile the transcriptomes of 180 highly purified HSCs (Bcl11a (+/+) and Bcl11a (-/-)). Detailed analysis of the RNA-seq data identifies cell cycle activity as the major source of transcriptomic variation in the HSC compartment, which allows reconstruction of HSC cell cycle progression in silico. Single-cell RNA-seq profiling of Bcl11a (-/-) HSCs reveals abnormal proliferative phenotypes. Analysis of lineage gene expression suggests that the Bcl11a (-/-) HSCs are constituted of two distinct myeloerythroid-restricted subpopulations. Remarkably, similar myeloid-restricted cells could also be detected in the wild-type HSC compartment, suggesting selective elimination of lymphoid-competent HSCs after Bcl11a deletion. These defects are experimentally validated in serial transplantation experiments where Bcl11a (-/-) HSCs are myeloerythroid-restricted and defective in self-renewal.
Conclusions: Our study demonstrates the power of single-cell transcriptomics in dissecting cellular process and lineage heterogeneities in stem cell compartments, and further reveals the molecular and cellular defects in the Bcl11a-deficient HSC compartment.
Figures





Similar articles
-
Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype.J Clin Invest. 2016 Oct 3;126(10):3868-3878. doi: 10.1172/JCI87885. Epub 2016 Sep 6. J Clin Invest. 2016. PMID: 27599293 Free PMC article.
-
Bcl11a Deficiency Leads to Hematopoietic Stem Cell Defects with an Aging-like Phenotype.Cell Rep. 2016 Sep 20;16(12):3181-3194. doi: 10.1016/j.celrep.2016.08.064. Cell Rep. 2016. PMID: 27653684 Free PMC article.
-
Bcl11a is essential for lymphoid development and negatively regulates p53.J Exp Med. 2012 Dec 17;209(13):2467-83. doi: 10.1084/jem.20121846. Epub 2012 Dec 10. J Exp Med. 2012. PMID: 23230003 Free PMC article.
-
In-vivo differentiation of adult hematopoietic stem cells from a single-cell point of view.Curr Opin Hematol. 2020 Jul;27(4):241-247. doi: 10.1097/MOH.0000000000000587. Curr Opin Hematol. 2020. PMID: 32398457 Review.
-
Differentiation-based model of hematopoietic stem cell functions and lineage pathways.Blood. 2018 Sep 13;132(11):1106-1113. doi: 10.1182/blood-2018-03-791517. Epub 2018 Jul 24. Blood. 2018. PMID: 30042097 Free PMC article. Review.
Cited by
-
Down-regulation of the transcriptional repressor ZNF802 (JAZF1) reactivates fetal hemoglobin in β0-thalassemia/HbE.Sci Rep. 2022 Mar 23;12(1):4952. doi: 10.1038/s41598-022-08920-8. Sci Rep. 2022. PMID: 35322124 Free PMC article.
-
A Small Key for a Heavy Door: Genetic Therapies for the Treatment of Hemoglobinopathies.Front Genome Ed. 2021 Feb 4;2:617780. doi: 10.3389/fgeed.2020.617780. eCollection 2020. Front Genome Ed. 2021. PMID: 34713239 Free PMC article. Review.
-
Single-cell genomics: coming of age.Genome Biol. 2016 May 10;17:97. doi: 10.1186/s13059-016-0960-x. Genome Biol. 2016. PMID: 27160975 Free PMC article. No abstract available.
-
Single-Cell Sequencing Technologies in Precision Oncology.Adv Exp Med Biol. 2022;1361:269-282. doi: 10.1007/978-3-030-91836-1_15. Adv Exp Med Biol. 2022. PMID: 35230694 Review.
-
BCL11A confers cell invasion and migration in androgen receptor-positive triple-negative breast cancer.Oncol Lett. 2020 Apr;19(4):2916-2924. doi: 10.3892/ol.2020.11383. Epub 2020 Feb 10. Oncol Lett. 2020. PMID: 32218847 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

