MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells
- PMID: 26387865
- PMCID: PMC4818967
- DOI: 10.1016/j.cmet.2015.09.003
MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells
Abstract
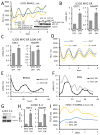
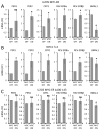
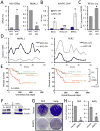
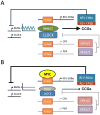
The MYC oncogene encodes MYC, a transcription factor that binds the genome through sites termed E-boxes (5'-CACGTG-3'), which are identical to the binding sites of the heterodimeric CLOCK-BMAL1 master circadian transcription factor. Hence, we hypothesized that ectopic MYC expression perturbs the clock by deregulating E-box-driven components of the circadian network in cancer cells. We report here that deregulated expression of MYC or N-MYC disrupts the molecular clock in vitro by directly inducing REV-ERBα to dampen expression and oscillation of BMAL1, and this could be rescued by knockdown of REV-ERB. REV-ERBα expression predicts poor clinical outcome for N-MYC-driven human neuroblastomas that have diminished BMAL1 expression, and re-expression of ectopic BMAL1 in neuroblastoma cell lines suppresses their clonogenicity. Further, ectopic MYC profoundly alters oscillation of glucose metabolism and perturbs glutaminolysis. Our results demonstrate an unsuspected link between oncogenic transformation and circadian and metabolic dysrhythmia, which we surmise to be advantageous for cancer.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Comment in
-
Metabolism: MYC clocks on.Nat Rev Cancer. 2015 Nov;15(11):636-7. doi: 10.1038/nrc4036. Epub 2015 Oct 15. Nat Rev Cancer. 2015. PMID: 26469145 No abstract available.
-
Stopping the Clock with MYC.Mol Cell. 2015 Nov 19;60(4):511-3. doi: 10.1016/j.molcel.2015.11.008. Mol Cell. 2015. PMID: 26590710
-
Correspondence: Reply to 'Oncogenic MYC persistently upregulates the molecular clock component REV-ERBα'.Nat Commun. 2017 Mar 23;8:14918. doi: 10.1038/ncomms14918. Nat Commun. 2017. PMID: 28332490 Free PMC article. No abstract available.
References
-
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. - PubMed
-
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. - PubMed
-
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK045586/DK/NIDDK NIH HHS/United States
- R01 CA057341/CA/NCI NIH HHS/United States
- F32 CA174148/CA/NCI NIH HHS/United States
- R01 CA123350/CA/NCI NIH HHS/United States
- RC1 MD004418/MD/NIMHD NIH HHS/United States
- R01 CA051497/CA/NCI NIH HHS/United States
- F32CA174148/CA/NCI NIH HHS/United States
- F32 CA180370/CA/NCI NIH HHS/United States
- R01CA057341/CA/NCI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- F32CA180370/CA/NCI NIH HHS/United States
- F30CA200347/CA/NCI NIH HHS/United States
- P01 CA104838/CA/NCI NIH HHS/United States
- F30 CA200347/CA/NCI NIH HHS/United States
- RC1MD004418/MD/NIMHD NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
- R01 NS054794/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

