Characterization of the cardiac succinylome and its role in ischemia-reperfusion injury
- PMID: 26388266
- PMCID: PMC4780049
- DOI: 10.1016/j.yjmcc.2015.09.005
Characterization of the cardiac succinylome and its role in ischemia-reperfusion injury
Abstract
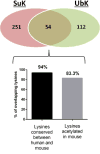
Succinylation refers to modification of lysine residues with succinyl groups donated by succinyl-CoA. Sirtuin5 (Sirt5) is a mitochondrial NAD(+)-dependent deacylase that catalyzes the removal of succinyl groups from proteins. Sirt5 and protein succinylation are conserved across species, suggesting functional importance of the modification. Sirt5 loss impacts liver metabolism but the role of succinylation in the heart has not been explored. We combined affinity enrichment with proteomics and mass spectrometry to analyze total succinylated lysine content of mitochondria isolated from WT and Sirt5(-/-) mouse hearts. We identified 887 succinylated lysine residues in 184 proteins. 44 peptides (5 proteins) occurred uniquely in WT samples, 289 (46 proteins) in Sirt5(-/-) samples, and 554 (133 proteins) were common to both groups. The 46 unique proteins in Sirt5(-/-) heart participate in metabolic processes such as fatty acid β-oxidation (Eci2) and branched chain amino acid catabolism, and include respiratory chain proteins (Ndufa7, 12, 13, Dhsa). We performed label-free analysis of the peptides common to WT and Sirt5(-/-) hearts. 16 peptides from 9 proteins were significantly increased in Sirt5(-/-) by at least 30%. The adenine nucleotide transporter 1 showed the highest increase in succinylation in Sirt5(-/-) (108.4 fold). The data indicate that succinylation is widespread in the heart and enriched in metabolic pathways. We examined whether the loss of Sirt5 would impact ischemia-reperfusion (I/R) injury and we found an increase in infarct size in Sirt5(-/-) hearts compared to WT littermates (68.5(+)/-1.1% Sirt5(-/-) vs 39.6(+)/(-) 6.8% WT) following 20min of ischemia and 90-min reperfusion. We further demonstrate that I/R injury in Sirt5(-/-) heart is restored to WT levels by pretreatment with dimethyl malonate, a competitive inhibitor of succinate dehydrogenase (SDH), implicating alteration in SDH activity as causative of the injury.
Keywords: Cardiac; Ischemia–reperfusion; Sirt5; Succinate; Succinylation.
Published by Elsevier Ltd.
Figures










Comment in
-
Not low hanging but still sweet: Metabolic proteomes in cardiovascular disease.J Mol Cell Cardiol. 2016 Jan;90:70-3. doi: 10.1016/j.yjmcc.2015.11.022. Epub 2015 Nov 22. J Mol Cell Cardiol. 2016. PMID: 26611885
References
-
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

