A comprehensive epigenome map of Plasmodium falciparum reveals unique mechanisms of transcriptional regulation and identifies H3K36me2 as a global mark of gene suppression
- PMID: 26388940
- PMCID: PMC4574195
- DOI: 10.1186/s13072-015-0029-1
A comprehensive epigenome map of Plasmodium falciparum reveals unique mechanisms of transcriptional regulation and identifies H3K36me2 as a global mark of gene suppression
Abstract
Background: Role of epigenetic mechanisms towards regulation of the complex life cycle/pathogenesis of Plasmodium falciparum, the causative agent of malaria, has been poorly understood. To elucidate stage-specific epigenetic regulation, we performed genome-wide mapping of multiple histone modifications of P. falciparum. Further to understand the differences in transcription regulation in P. falciparum and its host, human, we compared their histone modification profiles.
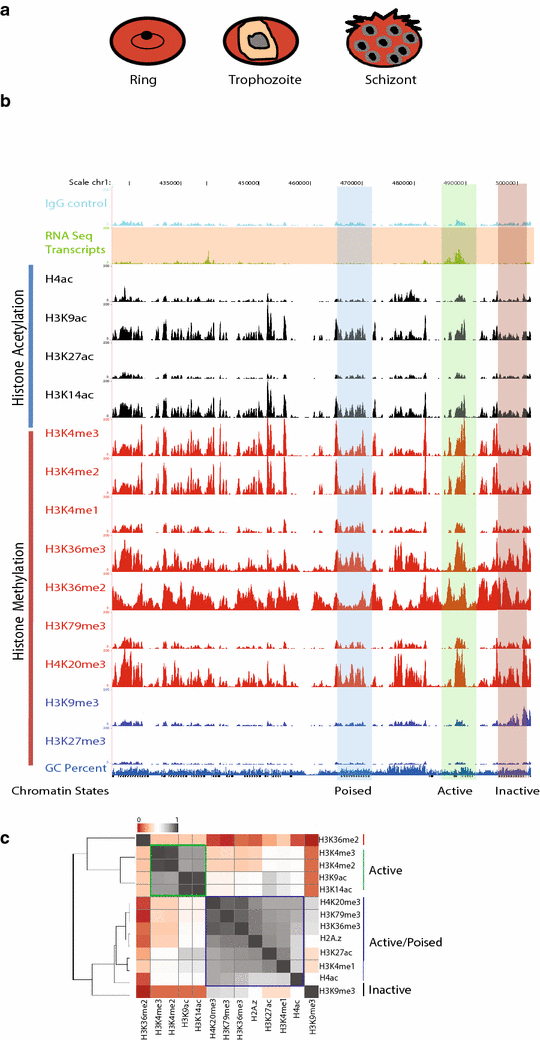
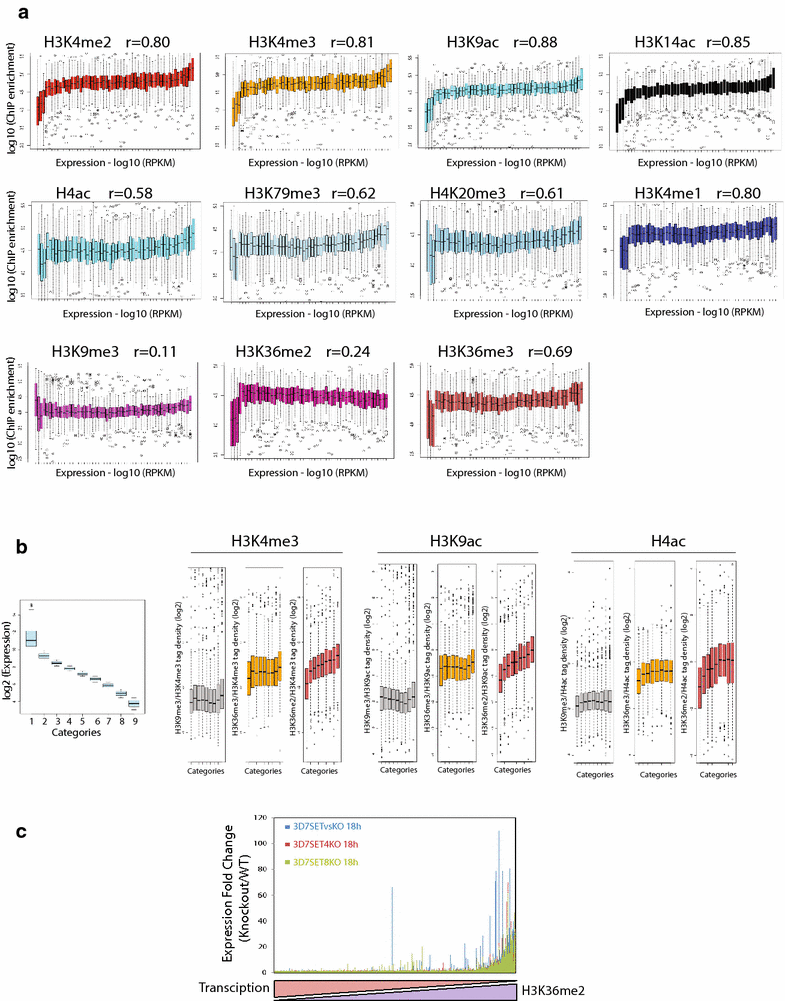
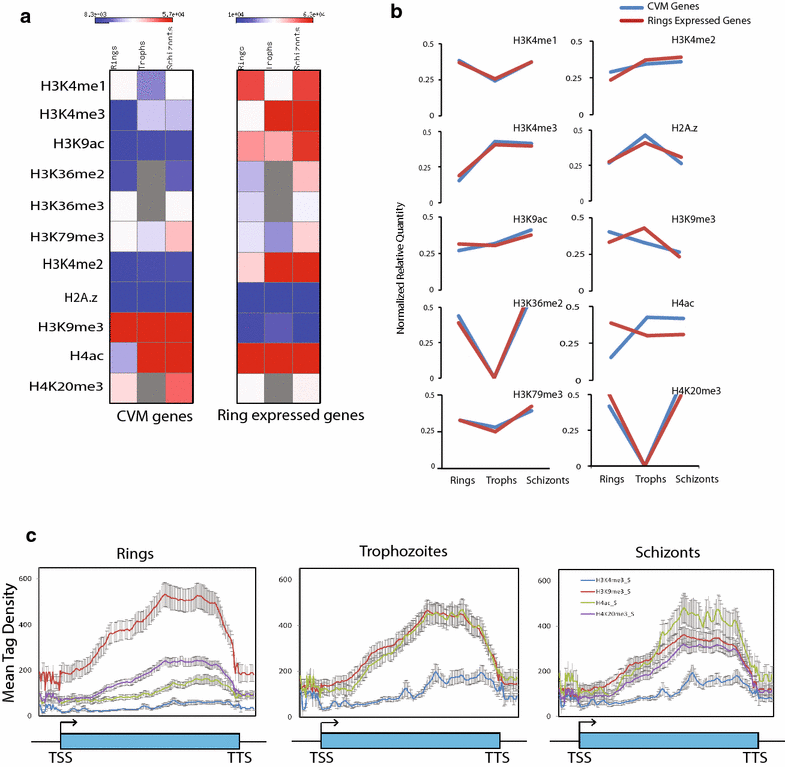
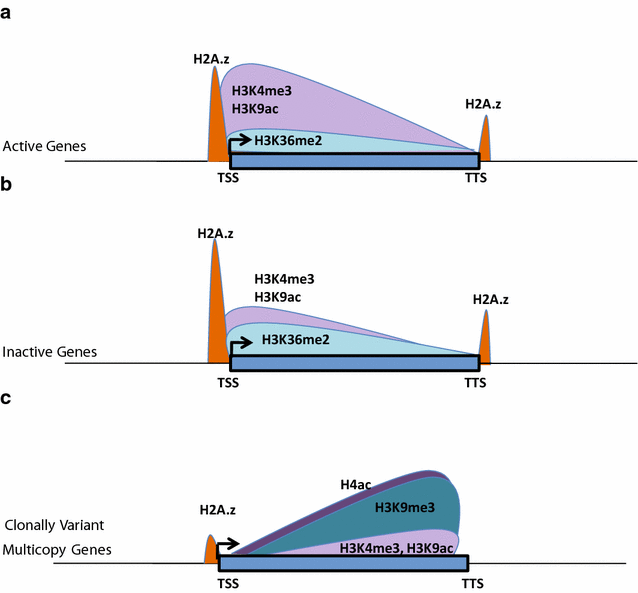
Results: Our comprehensive comparative analysis suggests distinct mode of transcriptional regulation in malaria parasite by virtue of poised genes and differential histone modifications. Furthermore, analysis of histone modification profiles predicted 562 genes producing anti-sense RNAs and 335 genes having bidirectional promoter activity, which raises the intriguing possibility of RNA-mediated regulation of transcription in P. falciparum. Interestingly, we found that H3K36me2 acts as a global repressive mark and gene regulation is fine tuned by the ratio of activation marks to H3K36me2 in P. falciparum. This novel mechanism of gene regulation is supported by the fact that knockout of SET genes (responsible for H3K36 methylation) leads to up-regulation of genes with highest occupancy of H3K36me2 in wild-type P. falciparum. Moreover, virulence (var) genes are mostly poised and marked by a unique set of activation (H4ac) and repression (H3K9me3) marks, which are mutually exclusive to other Plasmodium housekeeping genes.
Conclusions: Our study reveals unique plasticity in the epigenetic regulation in P. falciparum which can influence parasite virulence and pathogenicity. The observed differences in the histone code and transcriptional regulation in P. falciparum and its host will open new avenues for epigenetic drug development against malaria parasite.
Keywords: Chromatin; Genome-wide mapping; Histone modifications; Plasmodium; Transcription; Virulence and pathogenicity genes.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

