Tissue-specific expression of histone H3 variants diversified after species separation
- PMID: 26388943
- PMCID: PMC4574566
- DOI: 10.1186/s13072-015-0027-3
Tissue-specific expression of histone H3 variants diversified after species separation
Abstract
Background: The selective incorporation of appropriate histone variants into chromatin is critical for the regulation of genome function. Although many histone variants have been identified, a complete list has not been compiled.
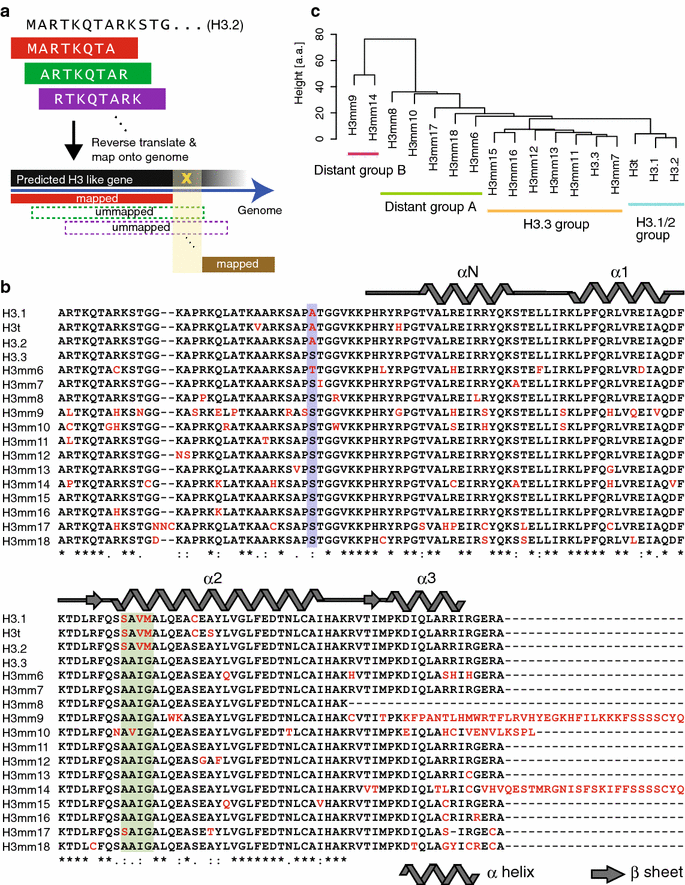
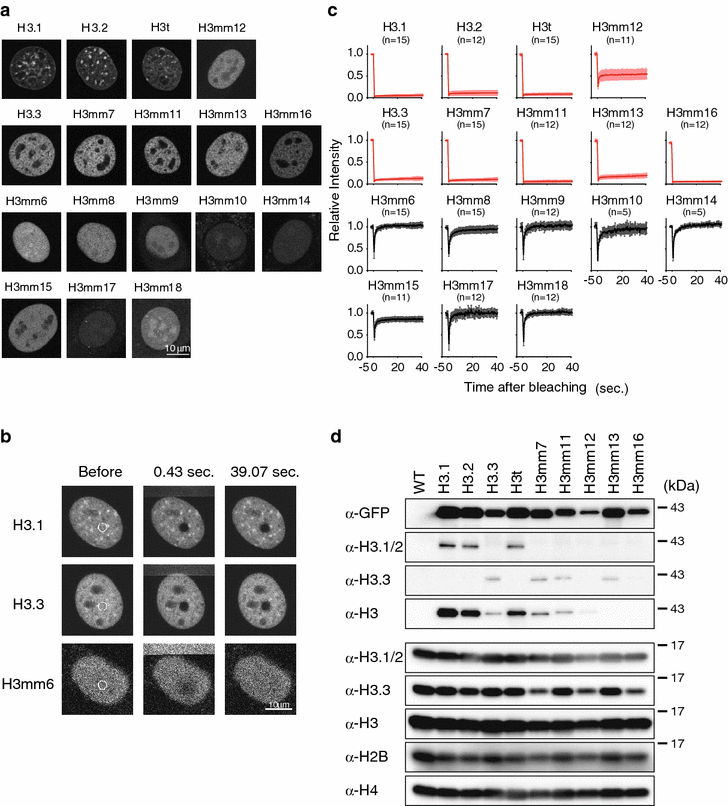
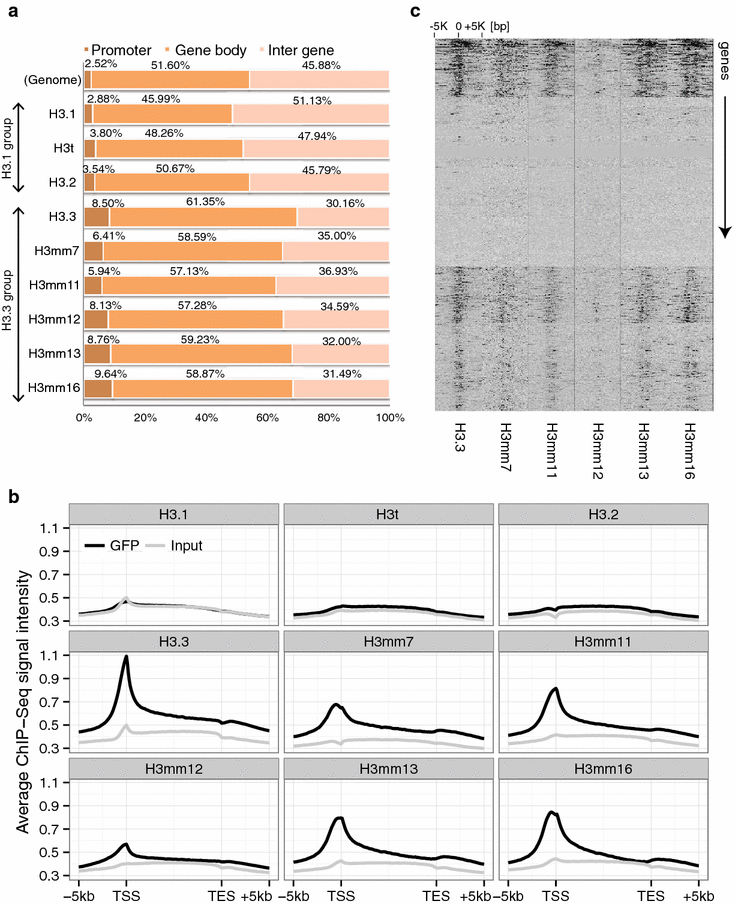
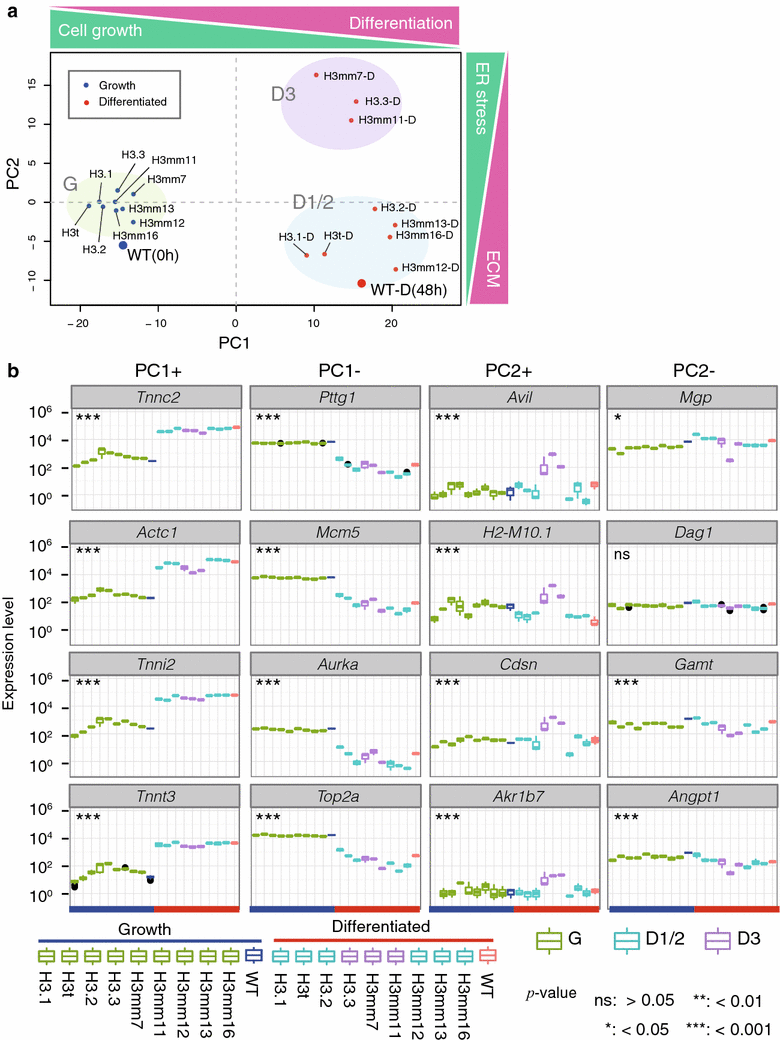
Results: We screened mouse, rat and human genomes by in silico hybridization using canonical histone sequences. In the mouse genome, we identified 14 uncharacterized H3 genes, among which 13 are similar to H3.3 and do not have human or rat counterparts, and one is similar to human testis-specific H3 variant, H3T/H3.4, and had a rat paralog. Although some of these genes were previously annotated as pseudogenes, their tissue-specific expression was confirmed by sequencing the 3'-UTR regions of the transcripts. Certain new variants were also detected at the protein level by mass spectrometry. When expressed as GFP-tagged versions in mouse C2C12 cells, some variants were stably incorporated into chromatin and the genome-wide distributions of most variants were similar to that of H3.3. Moreover, forced expression of H3 variants in chromatin resulted in alternate gene expression patterns after cell differentiation.
Conclusions: We comprehensively identified and characterized novel mouse H3 variant genes that encoded highly conserved amino acid sequences compared to known histone H3. We speculated that the diversity of H3 variants acquired after species separation played a role in regulating tissue-specific gene expression in individual species. Their biological relevance and evolutionary aspect involving pseudogene diversification will be addressed by further functional analysis.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

