ColE1-Plasmid Production in Escherichia coli: Mathematical Simulation and Experimental Validation
- PMID: 26389114
- PMCID: PMC4555960
- DOI: 10.3389/fbioe.2015.00127
ColE1-Plasmid Production in Escherichia coli: Mathematical Simulation and Experimental Validation
Abstract
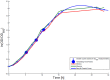
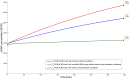
Plasmids have become very important as pharmaceutical gene vectors in the fields of gene therapy and genetic vaccination in the past years. In this study, we present a dynamic model to simulate the ColE1-like plasmid replication control, once for a DH5α-strain carrying a low copy plasmid (DH5α-pSUP 201-3) and once for a DH5α-strain carrying a high copy plasmid (DH5α-pCMV-lacZ) by using ordinary differential equations and the MATLAB software. The model includes the plasmid replication control by two regulatory RNA molecules (RNAI and RNAII) as well as the replication control by uncharged tRNA molecules. To validate the model, experimental data like RNAI- and RNAII concentration, plasmid copy number (PCN), and growth rate for three different time points in the exponential phase were determined. Depending on the sampled time point, the measured RNAI- and RNAII concentrations for DH5α-pSUP 201-3 reside between 6 ± 0.7 and 34 ± 7 RNAI molecules per cell and 0.44 ± 0.1 and 3 ± 0.9 RNAII molecules per cell. The determined PCNs averaged between 46 ± 26 and 48 ± 30 plasmids per cell. The experimentally determined data for DH5α-pCMV-lacZ reside between 345 ± 203 and 1086 ± 298 RNAI molecules per cell and 22 ± 2 and 75 ± 10 RNAII molecules per cell with an averaged PCN of 1514 ± 1301 and 5806 ± 4828 depending on the measured time point. As the model was shown to be consistent with the experimentally determined data, measured at three different time points within the growth of the same strain, we performed predictive simulations concerning the effect of uncharged tRNA molecules on the ColE1-like plasmid replication control. The hypothesis is that these tRNA molecules would have an enhancing effect on the plasmid production. The in silico analysis predicts that uncharged tRNA molecules would indeed increase the plasmid DNA production.
Keywords: biotechnology; high copy plasmid; modeling; ordinary differential equations; plasmid replication; small RNA; uncharged tRNA.
Figures


Similar articles
-
Replication regulation of ColE1-like plasmids in amino acid-starved Escherichia coli.Plasmid. 1998;39(1):48-62. doi: 10.1006/plas.1997.1319. Plasmid. 1998. PMID: 9473446
-
Maintenance of pBR322-derived plasmids without functional RNAI.Plasmid. 1991 Nov;26(3):186-200. doi: 10.1016/0147-619x(91)90042-u. Plasmid. 1991. PMID: 1721720
-
Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli.Mol Gen Genet. 1986 Apr;203(1):143-9. doi: 10.1007/BF00330395. Mol Gen Genet. 1986. PMID: 2423846
-
Processing of plasmid DNA with ColE1-like replication origin.Plasmid. 2004 May;51(3):149-61. doi: 10.1016/j.plasmid.2003.12.002. Plasmid. 2004. PMID: 15109822 Review.
-
Is tRNA only a translation factor or also a regulator of other processes?J Appl Genet. 2008;49(1):115-22. doi: 10.1007/BF03195257. J Appl Genet. 2008. PMID: 18263978 Review.
Cited by
-
Copy number variability of expression plasmids determined by cell sorting and Droplet Digital PCR.Microb Cell Fact. 2016 Dec 19;15(1):211. doi: 10.1186/s12934-016-0610-8. Microb Cell Fact. 2016. PMID: 27993152 Free PMC article.
-
Whole cell-based catalyst for enzymatic production of the osmolyte 2-O-α-glucosylglycerol.Microb Cell Fact. 2021 Apr 7;20(1):79. doi: 10.1186/s12934-021-01569-4. Microb Cell Fact. 2021. PMID: 33827582 Free PMC article.
-
Construction of an easy-to-use CRISPR-Cas9 system by patching a newly designed EXIT circuit.J Biol Eng. 2017 Sep 4;11:32. doi: 10.1186/s13036-017-0072-5. eCollection 2017. J Biol Eng. 2017. PMID: 28878819 Free PMC article.
References
-
- Alberts B., Johnson A., Walter P., Lewis J. (2007). Molecular Biology of the Cell, 5th Revised Edn New York & Abington: Garland Science, Taylor & Francis.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

