Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes
- PMID: 26389673
- PMCID: PMC4607110
- DOI: 10.1172/JCI82416
Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes
Abstract
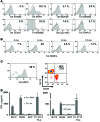
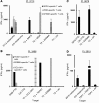
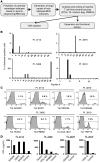
Adoptively transferred tumor-infiltrating T lymphocytes (TILs) that mediate complete regression of metastatic melanoma have been shown to recognize mutated epitopes expressed by autologous tumors. Here, in an attempt to develop a strategy for facilitating the isolation, expansion, and study of mutated antigen-specific T cells, we performed whole-exome sequencing on matched tumor and normal DNA isolated from 8 patients with metastatic melanoma. Candidate mutated epitopes were identified using a peptide-MHC-binding algorithm, and these epitopes were synthesized and used to generate panels of MHC tetramers that were evaluated for binding to tumor digests and cultured TILs used for the treatment of patients. This strategy resulted in the identification of 9 mutated epitopes from 5 of the 8 patients tested. Cells reactive with 8 of the 9 epitopes could be isolated from autologous peripheral blood, where they were detected at frequencies that were estimated to range between 0.4% and 0.002%. To the best of our knowledge, this represents the first demonstration of the successful isolation of mutation-reactive T cells from patients' peripheral blood prior to immune therapy, potentially providing the basis for designing personalized immunotherapies to treat patients with advanced cancer.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

