Autophagy signal transduction by ATG proteins: from hierarchies to networks
- PMID: 26390974
- PMCID: PMC4648967
- DOI: 10.1007/s00018-015-2034-8
Autophagy signal transduction by ATG proteins: from hierarchies to networks
Abstract
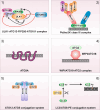
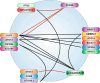
Autophagy represents an intracellular degradation process which is involved in both cellular homeostasis and disease settings. In the last two decades, the molecular machinery governing this process has been characterized in detail. To date, several key factors regulating this intracellular degradation process have been identified. The so-called autophagy-related (ATG) genes and proteins are central to this process. However, several additional molecules contribute to the outcome of an autophagic response. Several review articles describing the molecular process of autophagy have been published in the recent past. In this review article we would like to add the most recent findings to this knowledge, and to give an overview of the network character of the autophagy signaling machinery.
Keywords: ATG; Autophagy; LC3; PtdIns3K; ULK.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

