The NIDDK Information Network: A Community Portal for Finding Data, Materials, and Tools for Researchers Studying Diabetes, Digestive, and Kidney Diseases
- PMID: 26393351
- PMCID: PMC4578941
- DOI: 10.1371/journal.pone.0136206
The NIDDK Information Network: A Community Portal for Finding Data, Materials, and Tools for Researchers Studying Diabetes, Digestive, and Kidney Diseases
Abstract
The NIDDK Information Network (dkNET; http://dknet.org) was launched to serve the needs of basic and clinical investigators in metabolic, digestive and kidney disease by facilitating access to research resources that advance the mission of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). By research resources, we mean the multitude of data, software tools, materials, services, projects and organizations available to researchers in the public domain. Most of these are accessed via web-accessible databases or web portals, each developed, designed and maintained by numerous different projects, organizations and individuals. While many of the large government funded databases, maintained by agencies such as European Bioinformatics Institute and the National Center for Biotechnology Information, are well known to researchers, many more that have been developed by and for the biomedical research community are unknown or underutilized. At least part of the problem is the nature of dynamic databases, which are considered part of the "hidden" web, that is, content that is not easily accessed by search engines. dkNET was created specifically to address the challenge of connecting researchers to research resources via these types of community databases and web portals. dkNET functions as a "search engine for data", searching across millions of database records contained in hundreds of biomedical databases developed and maintained by independent projects around the world. A primary focus of dkNET are centers and projects specifically created to provide high quality data and resources to NIDDK researchers. Through the novel data ingest process used in dkNET, additional data sources can easily be incorporated, allowing it to scale with the growth of digital data and the needs of the dkNET community. Here, we provide an overview of the dkNET portal and its functions. We show how dkNET can be used to address a variety of use cases that involve searching for research resources.
Conflict of interest statement
Figures
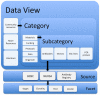



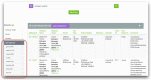


References
-
- BD2K Data Discovery Index Workshop Summary Report (2013) Available: http://bd2k.nih.gov/pdf/DDI_Workshop_summary.pdf.
-
- Cachat J, Bandrowski A, Grethe JS, Gupta A, Astakhov V, Imam F, et al.: A survey of the neuroscience resource landscape: perspectives from the neuroscience information framework. International review of neurobiology 2012, 103:39–68. - PubMed
-
- Gupta S, Bhatia KK. Deep questions in the”Deep or Hidden” web in B. V. Babu et al (eds) Proceedings of the Second International Conference on Soft Computing for Problem Solving, Dec 28030, 2012, Advances in Intelligent Systems and Computing 236 10.1007/978-81-322-1602-5_87.
-
- McKenna NJ, Howard CL, Aufiero M, Easton-Marks J, Steffen DL, Becnel LB, Magnuson MA, et al.: Research resource: dkCOIN, the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) consortium interconnectivity network: a pilot program to aggregate research resources generated by multiple research consortia. Molecular endocrinology (Baltimore, Md.) 2012, 26:1675–81. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

