Systematic Prediction of Scaffold Proteins Reveals New Design Principles in Scaffold-Mediated Signal Transduction
- PMID: 26393507
- PMCID: PMC4578958
- DOI: 10.1371/journal.pcbi.1004508
Systematic Prediction of Scaffold Proteins Reveals New Design Principles in Scaffold-Mediated Signal Transduction
Abstract
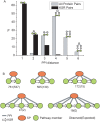
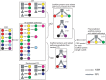
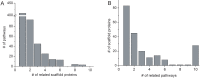
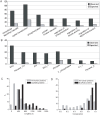
Scaffold proteins play a crucial role in facilitating signal transduction in eukaryotes by bringing together multiple signaling components. In this study, we performed a systematic analysis of scaffold proteins in signal transduction by integrating protein-protein interaction and kinase-substrate relationship networks. We predicted 212 scaffold proteins that are involved in 605 distinct signaling pathways. The computational prediction was validated using a protein microarray-based approach. The predicted scaffold proteins showed several interesting characteristics, as we expected from the functionality of scaffold proteins. We found that the scaffold proteins are likely to interact with each other, which is consistent with previous finding that scaffold proteins tend to form homodimers and heterodimers. Interestingly, a single scaffold protein can be involved in multiple signaling pathways by interacting with other scaffold protein partners. Furthermore, we propose two possible regulatory mechanisms by which the activity of scaffold proteins is coordinated with their associated pathways through phosphorylation process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- GM111514/GM/NIGMS NIH HHS/United States
- R01 GM076102/GM/NIGMS NIH HHS/United States
- U54 RR020839/RR/NCRR NIH HHS/United States
- U54 HG006434/HG/NHGRI NIH HHS/United States
- DP1 CA174423/CA/NCI NIH HHS/United States
- HG006434/HG/NHGRI NIH HHS/United States
- U24 CA160036/CA/NCI NIH HHS/United States
- CA174423/CA/NCI NIH HHS/United States
- RR020839/RR/NCRR NIH HHS/United States
- GM076102/GM/NIGMS NIH HHS/United States
- R01 GM111514/GM/NIGMS NIH HHS/United States
- R01 DK073368/DK/NIDDK NIH HHS/United States
- DK073368/DK/NIDDK NIH HHS/United States
- CA160036/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

