Mitochondrial Optic Atrophy (OPA) 1 Processing Is Altered in Response to Neonatal Hypoxic-Ischemic Brain Injury
- PMID: 26393574
- PMCID: PMC4613321
- DOI: 10.3390/ijms160922509
Mitochondrial Optic Atrophy (OPA) 1 Processing Is Altered in Response to Neonatal Hypoxic-Ischemic Brain Injury
Abstract
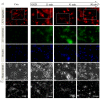
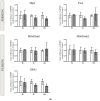
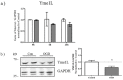
Perturbation of mitochondrial function and subsequent induction of cell death pathways are key hallmarks in neonatal hypoxic-ischemic (HI) injury, both in animal models and in term infants. Mitoprotective therapies therefore offer a new avenue for intervention for the babies who suffer life-long disabilities as a result of birth asphyxia. Here we show that after oxygen-glucose deprivation in primary neurons or in a mouse model of HI, mitochondrial protein homeostasis is altered, manifesting as a change in mitochondrial morphology and functional impairment. Furthermore we find that the mitochondrial fusion and cristae regulatory protein, OPA1, is aberrantly cleaved to shorter forms. OPA1 cleavage is normally regulated by a balanced action of the proteases Yme1L and Oma1. However, in primary neurons or after HI in vivo, protein expression of YmelL is also reduced, whereas no change is observed in Oma1 expression. Our data strongly suggest that alterations in mitochondria-shaping proteins are an early event in the pathogenesis of neonatal HI injury.
Keywords: OPA1; Oma1; Yme1L; hypoxia-ischaemia; mitochondria; neonatal brain injury; oxygen-glucose deprivation (OGD).
Figures





References
-
- Lawn J.E., Bahl R., Bergstrom S., Bhutta Z.A., Darmstadt G.L., Ellis M., English M., Kurinczuk J.J., Lee A.C., Merialdi M., et al. Setting research priorities to reduce almost one million deaths from birth asphyxia by 2015. PLoS Med. 2011;8:e1000389. doi: 10.1371/journal.pmed.1000389. - DOI - PMC - PubMed
-
- Azzopardi D., Wyatt J.S., Cady E.B., Delpy D.T., Baudin J., Stewart A.L., Hope P.L., Hamilton P.A., Reynolds E.O. Prognosis of newborn infants with hypoxic-ischemic brain injury assessed by phosphorus magnetic resonance spectroscopy. Pediatr. Res. 1989;25:445–451. doi: 10.1203/00006450-198905000-00004. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

