Enhanced virome sequencing using targeted sequence capture
- PMID: 26395152
- PMCID: PMC4665012
- DOI: 10.1101/gr.191049.115
Enhanced virome sequencing using targeted sequence capture
Abstract
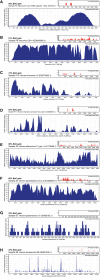
Metagenomic shotgun sequencing (MSS) is an important tool for characterizing viral populations. It is culture independent, requires no a priori knowledge of the viruses in the sample, and may provide useful genomic information. However, MSS can lack sensitivity and may yield insufficient data for detailed analysis. We have created a targeted sequence capture panel, ViroCap, designed to enrich nucleic acid from DNA and RNA viruses from 34 families that infect vertebrate hosts. A computational approach condensed ∼1 billion bp of viral reference sequence into <200 million bp of unique, representative sequence suitable for targeted sequence capture. We compared the effectiveness of detecting viruses in standard MSS versus MSS following targeted sequence capture. First, we analyzed two sets of samples, one derived from samples submitted to a diagnostic virology laboratory and one derived from samples collected in a study of fever in children. We detected 14 and 18 viruses in the two sets, comprising 19 genera from 10 families, with dramatic enhancement of genome representation following capture enrichment. The median fold-increases in percentage viral reads post-capture were 674 and 296. Median breadth of coverage increased from 2.1% to 83.2% post-capture in the first set and from 2.0% to 75.6% in the second set. Next, we analyzed samples containing a set of diverse anellovirus sequences and demonstrated that ViroCap could be used to detect viral sequences with up to 58% variation from the references used to select capture probes. ViroCap substantially enhances MSS for a comprehensive set of viruses and has utility for research and clinical applications.
© 2015 Wylie et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
