Large-scale live imaging of adult neural stem cells in their endogenous niche
- PMID: 26395477
- PMCID: PMC4631764
- DOI: 10.1242/dev.123018
Large-scale live imaging of adult neural stem cells in their endogenous niche
Abstract
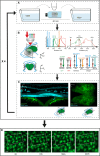
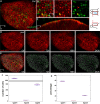
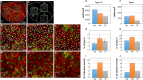
Live imaging of adult neural stem cells (aNSCs) in vivo is a technical challenge in the vertebrate brain. Here, we achieve long-term imaging of the adult zebrafish telencephalic neurogenic niche and track a population of >1000 aNSCs over weeks, by taking advantage of fish transparency at near-infrared wavelengths and of intrinsic multiphoton landmarks. This methodology enables us to describe the frequency, distribution and modes of aNSCs divisions across the entire germinal zone of the adult pallium, and to highlight regional differences in these parameters.
Keywords: Live imaging; Multiphoton microscopy; Neural stem cell; Pallium; SHG/THG; Zebrafish.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


References
-
- Chapouton P., Skupien P., Hesl B., Coolen M., Moore J. C., Madelaine R., Kremmer E., Faus-Kessler T., Blader P., Lawson N. D. et al. (2010). Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 30, 7961-7974. 10.1523/JNEUROSCI.6170-09.2010 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

