LRRK2 autophosphorylation enhances its GTPase activity
- PMID: 26396237
- PMCID: PMC4684519
- DOI: 10.1096/fj.15-277095
LRRK2 autophosphorylation enhances its GTPase activity
Abstract
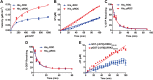
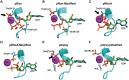
The leucine-rich repeat kinase (LRRK)-2 protein contains nonoverlapping GTPase and kinase domains, and mutation in either domain can cause Parkinson disease. GTPase proteins are critical upstream modulators of many effector protein kinases. In LRRK2, this paradigm may be reversed, as the kinase domain phosphorylates its own GTPase domain. In this study, we found that the ameba LRRK2 ortholog ROCO4 phosphorylates the GTPase domain [termed Ras-of-complex (ROC) domain in this family] of human LRRK2 on the same residues as the human LRRK2 kinase. Phosphorylation of ROC enhances its rate of GTP hydrolysis [from kcat (catalytic constant) 0.007 to 0.016 min(-1)], without affecting GTP or GDP dissociation kinetics [koff = 0.093 and 0.148 min(-1) for GTP and GDP, respectively). Phosphorylation also promotes the formation of ROC dimers, although GTPase activity appears to be equivalent between purified dimers and monomers. Modeling experiments show that phosphorylation induces conformational changes at the critical p-loop structure. Finally, ROC appears to be one of many GTPases phosphorylated in p-loop residues, as revealed by alignment of LRRK2 autophosphorylation sites with GTPases annotated in the phosphoproteome database. These results provide an example of a novel mechanism for kinase-mediated control of GTPase activity.
Keywords: G-protein; GTP-hydrolysis; Parkinson disease; ROC; phosphorylation.
© FASEB.
Figures



References
-
- Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. (2004) Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44, 595–600 - PubMed
-
- Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. (2004) Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44, 601–607 - PubMed
-
- Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J. M., Daniëls V., Lewis P., Jain S., Ding J., Syed A., Thomas K. J., Baekelandt V., Cookson M. R. (2008) The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 283, 16906–16914 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

