Virome Analysis of Transfusion Recipients Reveals a Novel Human Virus That Shares Genomic Features with Hepaciviruses and Pegiviruses
- PMID: 26396247
- PMCID: PMC4600124
- DOI: 10.1128/mBio.01466-15
Virome Analysis of Transfusion Recipients Reveals a Novel Human Virus That Shares Genomic Features with Hepaciviruses and Pegiviruses
Abstract
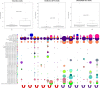
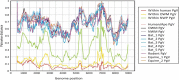
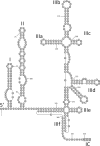
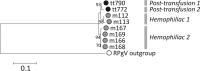
To investigate the transmission of novel infectious agents by blood transfusion, we studied changes in the virome composition of blood transfusion recipients pre- and posttransfusion. Using this approach, we detected and genetically characterized a novel human virus, human hepegivirus 1 (HHpgV-1), that shares features with hepatitis C virus (HCV) and human pegivirus (HPgV; formerly called GB virus C or hepatitis G virus). HCV and HPgV belong to the genera Hepacivirus and Pegivirus of the family Flaviviridae. HHpgV-1 was found in serum samples from two blood transfusion recipients and two hemophilia patients who had received plasma-derived clotting factor concentrates. In the former, the virus was detected only in the posttransfusion samples, indicating blood-borne transmission. Both hemophiliacs were persistently viremic over periods of at least 201 and 1,981 days. The 5' untranslated region (UTR) of HHpgV-1 contained a type IV internal ribosome entry site (IRES), structurally similar to although highly divergent in sequence from that of HCV and other hepaciviruses. However, phylogenetic analysis of nonstructural genes (NS3 and NS5B) showed that HHpgV-1 forms a branch within the pegivirus clade distinct from HPgV and homologs infecting other mammalian species. In common with some pegivirus variants infecting rodents and bats, the HHpgV-1 genome encodes a short, highly basic protein upstream of E1, potentially possessing a core-like function in packaging RNA during assembly. Identification of this new human virus, HHpgV-1, expands our knowledge of the range of genome configurations of these viruses and may lead to a reevaluation of the original criteria by which the genera Hepacivirus and Pegivirus are defined.
Importance: More than 30 million blood components are transfused annually in the United States alone. Surveillance for infectious agents in the blood supply is key to ensuring the safety of this critical resource for medicine and public health. Here, we report the identification of a new and highly diverse HCV/GB virus (GBV)-like virus from human serum samples. This new virus, human hepegivirus 1 (HHpgV-1), was found in serum samples from blood transfusion recipients, indicating its potential for transmission via transfusion products. We also found persistent long-term HHpgV-1 viremia in two hemophilia patients. HHpgV-1 is unique because it shares genetic similarity with both highly pathogenic HCV and the apparently nonpathogenic HPgV (GBV-C). Our results add to the list of human viruses and provide data to develop reagents to study virus transmission and disease association and for interrupting virus transmission and new human infections.
Copyright © 2015 Kapoor et al.
Figures


Comment in
-
New virus discovered in blood supply: Human hepegivirus-1 (HHpgV-1).Rev Argent Microbiol. 2016 Apr-Jun;48(2):180-1. doi: 10.1016/j.ram.2016.03.008. Epub 2016 Jun 3. Rev Argent Microbiol. 2016. PMID: 27268134 No abstract available.
References
-
- Ehmann WC, Eyster ME, Wilson SE, Andes WA, Goedert JJ. 1994. Relationship of CD4 lymphocyte counts to survival in a cohort of hemophiliacs infected with HIV. Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr 7:1095–1098. - PubMed
-
- Eyster ME, Fried MW, Di Bisceglie AM, Goedert JJ. 1994. Increasing hepatitis C virus RNA levels in hemophiliacs: relationship to human immunodeficiency virus infection and liver disease. Multicenter Hemophilia Cohort Study. Blood 84:1020–1023. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

