Proteomics of Xenopus development
- PMID: 26396253
- PMCID: PMC4767050
- DOI: 10.1093/molehr/gav052
Proteomics of Xenopus development
Abstract
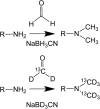
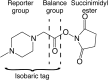
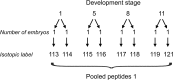
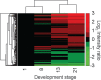
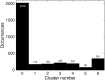
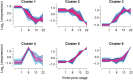
Modern mass spectrometry-based methods provide an exciting opportunity to characterize protein expression in the developing embryo. We have employed an isotopic labeling technology to quantify the expression dynamics of nearly 6000 proteins across six stages of development in Xenopus laevis from the single stage zygote through the mid-blastula transition and the onset of organogenesis. Approximately 40% of the proteins show significant changes in expression across the development stages. The expression changes for these proteins naturally falls into six clusters corresponding to major events that mark early Xenopus development. A subset of experiments in this study have quantified protein expression differences between single embryos at the same stage of development, showing that, within experimental error, embryos at the same developmental stage have identical protein expression levels.
Keywords: Xenopus laevis; early vertebrate development; expression clustering; iTRAQ chemistry; mass spectrometry; quantitative proteomics.
© The Author 2015. Published by Oxford University Press on behalf of the European Society of Human Reproduction and Embryology. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Aanes H, Collas P, Aleström P. Transcriptome dynamics and diversity in the early zebrafish embryo. Brief Funct Genomics 2014;13:95–105. - PubMed
-
- Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics 2002;1:947–955. - PubMed
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature 2003;422:198–207. - PubMed
-
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 2007;389:1017–1031. - PubMed
-
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 2009;4:484–494. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

