The Olfactory Transcriptome and Progression of Sexual Maturation in Homing Chum Salmon Oncorhynchus keta
- PMID: 26397372
- PMCID: PMC4580453
- DOI: 10.1371/journal.pone.0137404
The Olfactory Transcriptome and Progression of Sexual Maturation in Homing Chum Salmon Oncorhynchus keta
Abstract
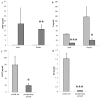
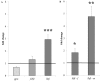
Reproductive homing migration of salmonids requires accurate interaction between the reception of external olfactory cues for navigation to the spawning grounds and the regulation of sexual maturation processes. This study aimed at providing insights into the hypothesized functional link between olfactory sensing of the spawning ground and final sexual maturation. We have therefore assessed the presence and expression levels of olfactory genes by RNA sequencing (RNAseq) of the olfactory rosettes in homing chum salmon Oncorhynchus keta Walbaum from the coastal sea to 75 km upstream the rivers at the pre-spawning ground. The progression of sexual maturation along the brain-pituitary-gonadal axis was assessed through determination of plasma steroid levels by time-resolved fluoroimmunoassays (TR-FIA), pituitary gonadotropin subunit expression and salmon gonadotropin-releasing hormone (sgnrh) expression in the brain by quantitative real-time PCR. RNAseq revealed the expression of 75 known and 27 unknown salmonid olfactory genes of which 13 genes were differentially expressed between fish from the pre-spawning area and from the coastal area, suggesting an important role of these genes in homing. A clear progression towards final maturation was characterised by higher plasma 17α,20β-dihydroxy-4-pregnen-3-one (DHP) levels, increased pituitary luteinizing hormone β subunit (lhβ) expression and sgnrh expression in the post brain, and lower plasma testosterone (T) and 17β-estradiol (E2) levels. Olfactomedins and ependymin are candidates among the differentially expressed genes that may connect olfactory reception to the expression of sgnrh to regulate final maturation.
Conflict of interest statement
Figures

References
-
- Wisby WJ, Hasler AD. Effect of olfactory occlusion on migrating silver salmon (Oncorhynchus kisutch). Journal of the Fisheries Research Board of Canada 1954; 11: 472–478.
-
- Ueda H. Artificial control of salmon homing migration and its application to salmon propagation. Bulletin of Tohoku National Fisheries Research Institute 1999; 62: 133–139.
-
- Urano A, Ando H, Ueda H. Molecular neuroendocrine basis of spawning migration in salmon In: Kwon HB, Joss JMP, Ishii S, editors. Recent progress in molecular and comparative endocrinology. Hormone Research Center, Kwangju; 1999. pp. 46–56.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

