Identification of microRNAs in the Toxigenic Dinoflagellate Alexandrium catenella by High-Throughput Illumina Sequencing and Bioinformatic Analysis
- PMID: 26398216
- PMCID: PMC4580472
- DOI: 10.1371/journal.pone.0138709
Identification of microRNAs in the Toxigenic Dinoflagellate Alexandrium catenella by High-Throughput Illumina Sequencing and Bioinformatic Analysis
Abstract
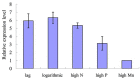
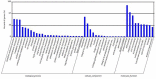
Micro-ribonucleic acids (miRNAs) are a large group of endogenous, tiny, non-coding RNAs consisting of 19-25 nucleotides that regulate gene expression at either the transcriptional or post-transcriptional level by mediating gene silencing in eukaryotes. They are considered to be important regulators that affect growth, development, and response to various stresses in plants. Alexandrium catenella is an important marine toxic phytoplankton species that can cause harmful algal blooms (HABs). To date, identification and function analysis of miRNAs in A. catenella remain largely unexamined. In this study, high-throughput sequencing was performed on A. catenella to identify and quantitatively profile the repertoire of small RNAs from two different growth phases. A total of 38,092,056 and 32,969,156 raw reads were obtained from the two small RNA libraries, respectively. In total, 88 mature miRNAs belonging to 32 miRNA families were identified. Significant differences were found in the member number, expression level of various families, and expression abundance of each member within a family. A total of 15 potentially novel miRNAs were identified. Comparative profiling showed that 12 known miRNAs exhibited differential expression between the lag phase and the logarithmic phase. Real-time quantitative RT-PCR (qPCR) was performed to confirm the expression of two differentially expressed miRNAs that were one up-regulated novel miRNA (aca-miR-3p-456915), and one down-regulated conserved miRNA (tae-miR159a). The expression trend of the qPCR assay was generally consistent with the deep sequencing result. Target predictions of the 12 differentially expressed miRNAs resulted in 1813 target genes. Gene ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes pathway database (KEGG) annotations revealed that some miRNAs were associated with growth and developmental processes of the alga. These results provide insights into the roles that miRNAs play in the growth of A. catenella, and they provide the basis for further studies of the molecular mechanisms that underlie bloom growth in red tides species.
Conflict of interest statement
Figures



Similar articles
-
Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing.BMC Genomics. 2015 Oct 22;16:835. doi: 10.1186/s12864-015-2010-6. BMC Genomics. 2015. PMID: 26490136 Free PMC article.
-
Identification and characterization of immune-related microRNAs in blunt snout bream, Megalobrama amblycephala.Fish Shellfish Immunol. 2016 Feb;49:470-92. doi: 10.1016/j.fsi.2015.12.013. Epub 2016 Jan 7. Fish Shellfish Immunol. 2016. PMID: 26773859
-
Identification of miRNAs and their targets through high-throughput sequencing and degradome analysis in male and female Asparagus officinalis.BMC Plant Biol. 2016 Apr 12;16:80. doi: 10.1186/s12870-016-0770-z. BMC Plant Biol. 2016. PMID: 27068118 Free PMC article.
-
Exploring dinoflagellate biology with high-throughput proteomics.Harmful Algae. 2018 May;75:16-26. doi: 10.1016/j.hal.2018.03.010. Epub 2018 Apr 16. Harmful Algae. 2018. PMID: 29778222 Review.
-
MicroRNA profiling: approaches and considerations.Nat Rev Genet. 2012 Apr 18;13(5):358-69. doi: 10.1038/nrg3198. Nat Rev Genet. 2012. PMID: 22510765 Free PMC article. Review.
Cited by
-
Distinctive Nuclear Features of Dinoflagellates with A Particular Focus on Histone and Histone-Replacement Proteins.Microorganisms. 2018 Dec 14;6(4):128. doi: 10.3390/microorganisms6040128. Microorganisms. 2018. PMID: 30558155 Free PMC article. Review.
-
Calmodulin and Its Interactive Proteins Participate in Regulating the Explosive Growth of Alexandrium pacificum (Dinoflagellate).Int J Mol Sci. 2021 Dec 23;23(1):145. doi: 10.3390/ijms23010145. Int J Mol Sci. 2021. PMID: 35008568 Free PMC article.
-
Identification of microRNAs Actively Involved in Fatty Acid Biosynthesis in Developing Brassica napus Seeds Using High-Throughput Sequencing.Front Plant Sci. 2016 Oct 24;7:1570. doi: 10.3389/fpls.2016.01570. eCollection 2016. Front Plant Sci. 2016. PMID: 27822220 Free PMC article.
-
The Genetic Basis of Toxin Biosynthesis in Dinoflagellates.Microorganisms. 2019 Jul 29;7(8):222. doi: 10.3390/microorganisms7080222. Microorganisms. 2019. PMID: 31362398 Free PMC article. Review.
-
Integrated omics unveil the secondary metabolic landscape of a basal dinoflagellate.BMC Biol. 2020 Oct 13;18(1):139. doi: 10.1186/s12915-020-00873-6. BMC Biol. 2020. PMID: 33050904 Free PMC article.
References
-
- Sournia A. Red tide and toxic marine phytoplankton of the world ocean: an inquiry into biodiversity Lavoisier, Paris(France) 1995; 103–112.
-
- Stoecker DK. Mixotrophy among Dinoflagellates1.Journal of Eukaryotic Microbiology. 1999;46: 397–401.
-
- Burkholder JM, Glibert PM and Skelton HM. Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae. 2008;8: 77–93.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

