A gene expression resource generated by genome-wide lacZ profiling in the mouse
- PMID: 26398943
- PMCID: PMC4631787
- DOI: 10.1242/dmm.021238
A gene expression resource generated by genome-wide lacZ profiling in the mouse
Abstract
Knowledge of the expression profile of a gene is a critical piece of information required to build an understanding of the normal and essential functions of that gene and any role it may play in the development or progression of disease. High-throughput, large-scale efforts are on-going internationally to characterise reporter-tagged knockout mouse lines. As part of that effort, we report an open access adult mouse expression resource, in which the expression profile of 424 genes has been assessed in up to 47 different organs, tissues and sub-structures using a lacZ reporter gene. Many specific and informative expression patterns were noted. Expression was most commonly observed in the testis and brain and was most restricted in white adipose tissue and mammary gland. Over half of the assessed genes presented with an absent or localised expression pattern (categorised as 0-10 positive structures). A link between complexity of expression profile and viability of homozygous null animals was observed; inactivation of genes expressed in ≥ 21 structures was more likely to result in reduced viability by postnatal day 14 compared with more restricted expression profiles. For validation purposes, this mouse expression resource was compared with Bgee, a federated composite of RNA-based expression data sets. Strong agreement was observed, indicating a high degree of specificity in our data. Furthermore, there were 1207 observations of expression of a particular gene in an anatomical structure where Bgee had no data, indicating a large amount of novelty in our data set. Examples of expression data corroborating and extending genotype-phenotype associations and supporting disease gene candidacy are presented to demonstrate the potential of this powerful resource.
Keywords: Gene expression; Mouse; Resource; lacZ reporter.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


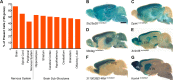


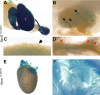

Comment in
-
Into the blue: the importance of murine lacZ gene expression profiling in understanding and treating human disease.Dis Model Mech. 2015 Nov;8(11):1341-3. doi: 10.1242/dmm.023606. Dis Model Mech. 2015. PMID: 26512121 Free PMC article.
References
-
- Adams N. C. and Gale N. W. (2006). High resolution gene expression analysis in mice using genetically inserted reporter genes. In Principle and Practice Mammalian and Avian Transgenesis - New Approaches (ed. Pease S. and Lois C.), pp. 132-172. Springer.
-
- Adissu H. A., Estabel J., Sunter D., Tuck E., Hooks Y., Carragher D. M., Clarke K., Karp N. A., Sanger Mouse Genetics Project, Newbigging S. et al. (2014). Histopathology reveals correlative and unique phenotypes in a high-throughput mouse phenotyping screen. Dis. Model. Mech. 7, 515-524. 10.1242/dmm.015263 - DOI - PMC - PubMed
-
- Bassett J. H. D., Gogakos A., White J. K., Evans H., Jacques R. M., van der Spek A. H., Sanger Mouse Genetics Project, Ramirez-Solis R., Ryder E., Sunter D. et al. (2012). Rapid-throughput skeletal phenotyping of 100 knockout mice identifies 9 new genes that determine bone strength. PLoS Genet. 8, e1002858 10.1371/journal.pgen.1002858 - DOI - PMC - PubMed
-
- Bastian F., Parmentier G., Roux J., Moretti S., Laudet V. and Robinson-Rechavi M. (2008). Bgee: integrating and comparing heterogeneous transcriptome data among species. In Data Integration in the Life Sciences (ed. Bairoch, Amos; Cohen-Boulakia, Sarah; Froidevaux, Christine), pp. 124-131. Berlin; Heidelberg: Springer.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

