Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes
- PMID: 26399631
- PMCID: PMC4654747
- DOI: 10.1007/s00401-015-1478-0
Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes
Abstract
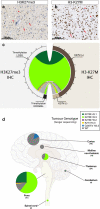
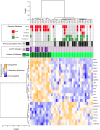
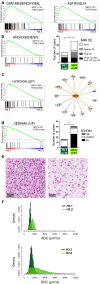
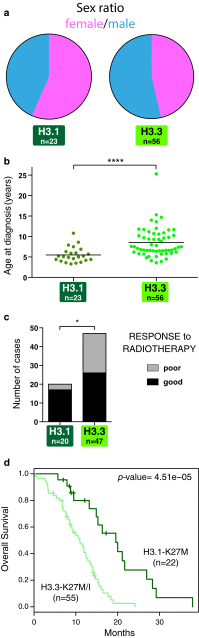
Diffuse intrinsic pontine glioma (DIPG) is the most severe paediatric solid tumour, with no significant therapeutic progress made in the past 50 years. Recent studies suggest that diffuse midline glioma, H3-K27M mutant, may comprise more than one biological entity. The aim of the study was to determine the clinical and biological variables that most impact their prognosis. Ninety-one patients with classically defined DIPG underwent a systematic stereotactic biopsy and were included in this observational retrospective study. Histone H3 genes mutations were assessed by immunochemistry and direct sequencing, whilst global gene expression profiling and chromosomal imbalances were determined by microarrays. A full description of the MRI findings at diagnosis and at relapse was integrated with the molecular profiling data and clinical outcome. All DIPG but one were found to harbour either a somatic H3-K27M mutation and/or loss of H3K27 trimethylation. We also discovered a novel K27M mutation in HIST2H3C, and a lysine-to-isoleucine substitution (K27I) in H3F3A, also creating a loss of trimethylation. Patients with tumours harbouring a K27M mutation in H3.3 (H3F3A) did not respond clinically to radiotherapy as well, relapsed significantly earlier and exhibited more metastatic recurrences than those in H3.1 (HIST1H3B/C). H3.3-K27M-mutated DIPG have a proneural/oligodendroglial phenotype and a pro-metastatic gene expression signature with PDGFRA activation, while H3.1-K27M-mutated tumours exhibit a mesenchymal/astrocytic phenotype and a pro-angiogenic/hypoxic signature supported by expression profiling and radiological findings. H3K27 alterations appear as the founding event in DIPG and the mutations in the two main histone H3 variants drive two distinct oncogenic programmes with potential specific therapeutic targets.
Figures

Comment in
-
Commentary on "Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes".Acta Neuropathol. 2016 May;131(5):793-4. doi: 10.1007/s00401-016-1567-8. Epub 2016 Mar 30. Acta Neuropathol. 2016. PMID: 27026412 No abstract available.
-
Histone H3 genotyping refines clinico-radiological diagnostic and prognostic criteria in DIPG.Acta Neuropathol. 2016 May;131(5):795-6. doi: 10.1007/s00401-016-1568-7. Epub 2016 Apr 1. Acta Neuropathol. 2016. PMID: 27038188 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

