Transcriptome-enabled marker discovery and mapping of plastochron-related genes in Petunia spp
- PMID: 26400485
- PMCID: PMC4581106
- DOI: 10.1186/s12864-015-1931-4
Transcriptome-enabled marker discovery and mapping of plastochron-related genes in Petunia spp
Abstract
Background: Petunia (Petunia × hybrida), derived from a hybrid between P. axillaris and P. integrifolia, is one of the most economically important bedding plant crops and Petunia spp. serve as model systems for investigating the mechanisms underlying diverse mating systems and pollination syndromes. In addition, we have previously described genetic variation and quantitative trait loci (QTL) related to petunia development rate and morphology, which represent important breeding targets for the floriculture industry to improve crop production and performance. Despite the importance of petunia as a crop, the floriculture industry has been slow to adopt marker assisted selection to facilitate breeding strategies and there remains a limited availability of sequences and molecular markers from the genus compared to other economically important members of the Solanaceae family such as tomato, potato and pepper.
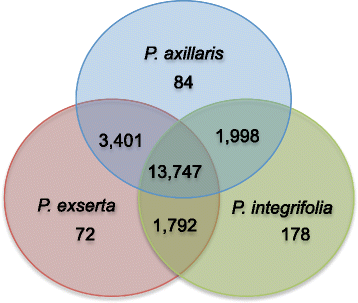
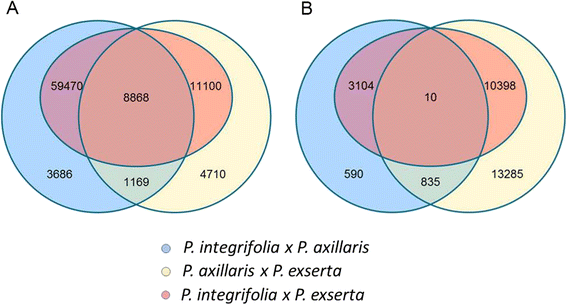
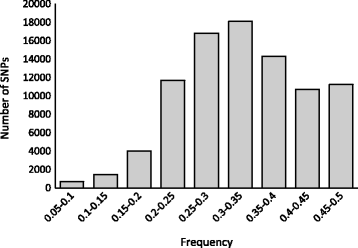
Results: Here we report the de novo assembly, annotation and characterization of transcriptomes from P. axillaris, P. exserta and P. integrifolia. Each transcriptome assembly was derived from five tissue libraries (callus, 3-week old seedlings, shoot apices, flowers of mixed developmental stages, and trichomes). A total of 74,573, 54,913, and 104,739 assembled transcripts were recovered from P. axillaris, P. exserta and P. integrifolia, respectively and following removal of multiple isoforms, 32,994 P. axillaris, 30,225 P. exserta, and 33,540 P. integrifolia high quality representative transcripts were extracted for annotation and expression analysis. The transcriptome data was mined for single nucleotide polymorphisms (SNP) and simple sequence repeat (SSR) markers, yielding 89,007 high quality SNPs and 2949 SSRs, respectively. 15,701 SNPs were computationally converted into user-friendly cleaved amplified polymorphic sequence (CAPS) markers and a subset of SNP and CAPS markers were experimentally verified. CAPS markers developed from plastochron-related homologous transcripts from P. axillaris were mapped in an interspecific Petunia population and evaluated for co-localization with QTL for development rate.
Conclusions: The high quality of the three Petunia spp. transcriptomes coupled with the utility of the SNP data will serve as a resource for further exploration of genetic diversity within the genus and will facilitate efforts to develop genetic and physical maps to aid the identification of QTL associated with traits of interest.
Figures



References
-
- Stehmann JR, Lorenz-Lemke AP, Freitas LB, Semir J. The genus Petunia. In: Gerats T, Strommer J, editors. Petunia Evolutionary, Developmental and Physiological Genetics. New York: Springer; 2009. pp. 1–28.
-
- Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J. 2012;70(5):739–49. doi: 10.1111/j.1365-313X.2012.04908.x. - DOI - PubMed
-
- Kelly RO, Deng ZA, Harbaugh BK. Evaluation of 125 petunia cultivars as bedding plants and establishment of class standards. Horttechnology. 2007;17(3):386–96.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

