Infectious Bronchitis Coronavirus Inhibits STAT1 Signaling and Requires Accessory Proteins for Resistance to Type I Interferon Activity
- PMID: 26401035
- PMCID: PMC4645315
- DOI: 10.1128/JVI.01057-15
Infectious Bronchitis Coronavirus Inhibits STAT1 Signaling and Requires Accessory Proteins for Resistance to Type I Interferon Activity
Abstract
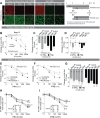
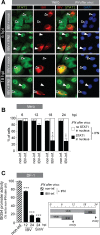
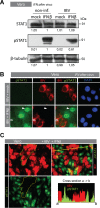
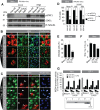
The innate immune response is the first line of defense against viruses, and type I interferon (IFN) is a critical component of this response. Similar to other viruses, the gammacoronavirus infectious bronchitis virus (IBV) has evolved under evolutionary pressure to evade and counteract the IFN response to enable its survival. Previously, we reported that IBV induces a delayed activation of the IFN response. In the present work, we describe the resistance of IBV to IFN and the potential role of accessory proteins herein. We show that IBV is fairly resistant to the antiviral state induced by IFN and identify that viral accessory protein 3a is involved in resistance to IFN, as its absence renders IBV less resistant to IFN treatment. In addition to this, we found that independently of its accessory proteins, IBV inhibits IFN-mediated phosphorylation and translocation of STAT1. In summary, we show that IBV uses multiple strategies to counteract the IFN response.
Importance: In the present study, we show that infectious bronchitis virus (IBV) is resistant to IFN treatment and identify a role for accessory protein 3a in the resistance against the type I IFN response. We also demonstrate that, in a time-dependent manner, IBV effectively interferes with IFN signaling and that its accessory proteins are dispensable for this activity. This study demonstrates that the gammacoronavirus IBV, similar to its mammalian counterparts, has evolved multiple strategies to efficiently counteract the IFN response of its avian host, and it identifies accessory protein 3a as multifaceted antagonist of the avian IFN system.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Britton P, Cavanagh D. 2007. Avian coronavirus diseases and infectious bronchitis vaccine development, p 161–181. In Thiel V. (ed), Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

