Genetic Diversity within Alphaherpesviruses: Characterization of a Novel Variant of Herpes Simplex Virus 2
- PMID: 26401046
- PMCID: PMC4665239
- DOI: 10.1128/JVI.01959-15
Genetic Diversity within Alphaherpesviruses: Characterization of a Novel Variant of Herpes Simplex Virus 2
Abstract
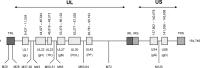
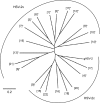
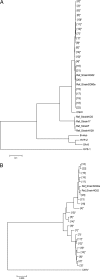
Very low levels of variability have been reported for the herpes simplex virus 2 (HSV-2) genome. We recently described a new genetic variant of HSV-2 (HSV-2v) characterized by a much higher degree of variability for the UL30 gene (DNA polymerase) than observed for the HG52 reference strain. Retrospective screening of 505 clinical isolates of HSV-2 by a specific real-time PCR assay targeting the UL30 gene led to the identification of 13 additional HSV-2v isolates, resulting in an overall prevalence of 2.8%. Phylogenetic analyses on the basis of microsatellite markers and gene sequences showed clear differences between HSV-2v and classical HSV-2. Thirteen of the 14 patients infected with HSV-2v originated from West or Central Africa, and 9 of these patients were coinfected with HIV. These results raise questions about the origin of this new virus. Preliminary results suggest that HSV-2v may have acquired genomic segments from chimpanzee alphaherpesvirus (ChHV) by recombination.
Importance: This article deals with the highly topical question of the origin of this new HSV-2 variant identified in patients with HIV coinfection originating mostly from West or Central Africa. HSV-2v clearly differed from classical HSV-2 isolates in phylogenetic analyses and may be linked to simian ChHV. This new HSV-2 variant highlights the possible occurrence of recombination between human and simian herpesviruses under natural conditions, potentially presenting greater challenges for the future.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
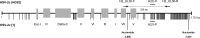


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

