Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus
- PMID: 26401053
- PMCID: PMC4654050
- DOI: 10.1093/hmg/ddv397
Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus
Abstract
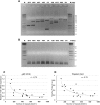
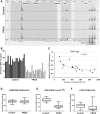
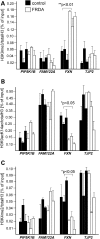
Friedreich's ataxia (FRDA) is a severe neurodegenerative disease caused by homozygous expansion of the guanine-adenine-adenine (GAA) repeats in intron 1 of the FXN gene leading to transcriptional repression of frataxin expression. Post-translational histone modifications that typify heterochromatin are enriched in the vicinity of the repeats, whereas active chromatin marks in this region are underrepresented in FRDA samples. Yet, the immediate effect of the expanded repeats on transcription progression through FXN and their long-range effect on the surrounding genomic context are two critical questions that remain unanswered in the molecular pathogenesis of FRDA. To address these questions, we conducted next-generation RNA sequencing of a large cohort of FRDA and control primary fibroblasts. This comprehensive analysis revealed that the GAA-induced silencing effect does not influence expression of neighboring genes upstream or downstream of FXN. Furthermore, no long-range silencing effects were detected across a large portion of chromosome 9. Additionally, results of chromatin immunoprecipitation studies confirmed that histone modifications associated with repressed transcription are confined to the FXN locus. Finally, deep sequencing of FXN pre-mRNA molecules revealed a pronounced defect in the transcription elongation rate in FRDA cells when compared with controls. These results indicate that approaches aimed to reactivate frataxin expression should simultaneously address deficits in transcription initiation and elongation at the FXN locus.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Pandolfo M. (2006) In Wells R.D., Ashizawa T. (eds), Genetic Instabilities and Neurological Diseases. Elsevier-Academic Press, San Diego, CA, pp. 277–296.
-
- Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A. et al. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423–1427. - PubMed
-
- Montermini L., Andermann E., Labuda M., Richter A., Pandolfo M., Cavalcanti F., Pianese L., Iodice L., Farina G., Monticelli A. et al. (1997) The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet., 6, 1261–1266. - PubMed
-
- Martelli A., Wattenhofer-Donze M., Schmucker S., Bouvet S., Reutenauer L., Puccio H. (2007) Frataxin is essential for extramitochondrial Fe–S cluster proteins in mammalian tissues. Hum. Mol. Genet., 16, 2651–2658. - PubMed
-
- Bulteau A.L., O'Neill H.A., Kennedy M.C., Ikeda-Saito M., Isaya G., Szweda L.I. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science, 305, 242–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

